2AX2
 
 | | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Budayova-Spano, M, Fisher, S.Z, Dauvergne, M.T, Silverman, D.N, Myles, D.A.A, McKenna, R.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3SS2
 
 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8DHD
 
 | | Neutron crystal structure of maltotetraose bound tmMBP | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S, Myles, D.A. | | Deposit date: | 2022-06-27 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Mapping periplasmic binding protein oligosaccharide recognition with neutron crystallography.
Sci Rep, 12, 2022
|
|
6U0E
 
 | |
6U0F
 
 | |
6U0B
 
 | |
6U0C
 
 | |
1LJ7
 
 | | Crystal structure of calcium-depleted human C-reactive protein from perfectly twinned data | | Descriptor: | C-reactive protein | | Authors: | Ramadan, M.A, Shrive, A.K, Holden, D, Myles, D.A, Volanakis, J.E, DeLucas, L.J, Greenhough, T.J. | | Deposit date: | 2002-04-19 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The three-dimensional structure of calcium-depleted human C-reactive protein from perfectly twinned crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8VJ0
 
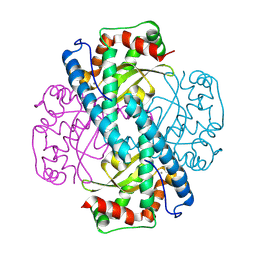 | | Neutron Structure of Oxidized Trp161Phe MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Nat Commun, 15, 2024
|
|
8VJ4
 
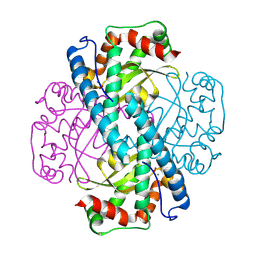 | | X-ray Counterpart to the Neutron Structure of Peroxide-Soaked Trp161Phe MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Nat Commun, 15, 2024
|
|
8VJ8
 
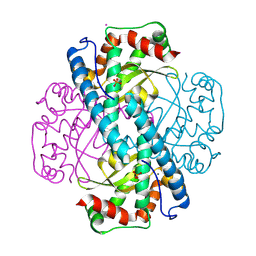 | | X-ray Counterpart to Neutron Structure of Reduced Trp161Phe MnSOD | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Nat Commun, 15, 2024
|
|
8VHW
 
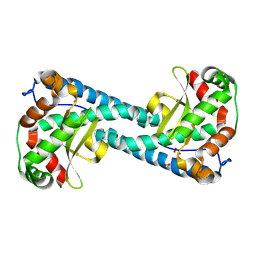 | | Neutron Structure of Peroxide-Soaked Trp161Phe MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Nat Commun, 15, 2024
|
|
8VJ5
 
 | | Peroxide-Soaked MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Nat Commun, 15, 2024
|
|
8VHY
 
 | | Neutron Structure of Reduced Trp161Phe MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Nat Commun, 15, 2024
|
|
5HM4
 
 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | Descriptor: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | Authors: | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
5H8Z
 
 | |
1OEX
 
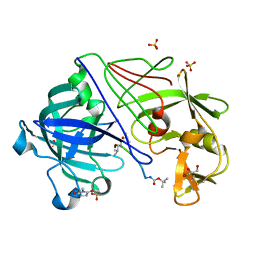 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OEW
 
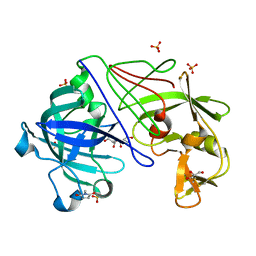 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1YRD
 
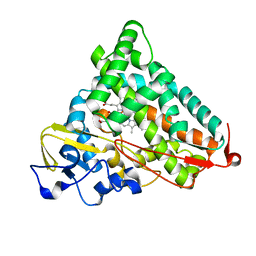 | | X-ray crystal structure of PERDEUTERATED Cytochrome P450cam | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Meilleur, F, Dauvergne, M.-T, Schlichting, I, Myles, D.A.A. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Production and X-ray crystallographic analysis of fully deuterated cytochrome P450cam.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YRC
 
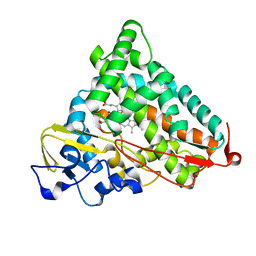 | | X-ray Crystal Structure of hydrogenated Cytochrome P450cam | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Meilleur, F, Dauvergne, M.-T, Schlichting, I, Myles, D.A.A. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Production and X-ray crystallographic analysis of fully deuterated cytochrome P450cam.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6NAF
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Selvaraj, B, Park, J, Cuneo, M.J, Myles, D.A.A, Baker, D. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.923 Å), X-RAY DIFFRACTION | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
3RZ6
 
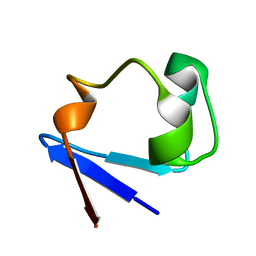 | | Neutron structure of perdeuterated rubredoxin using 40 hours 1st pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RZT
 
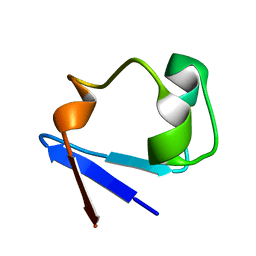 | | Neutron structure of perdeuterated rubredoxin using rapid (14 hours) data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.7504 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RYG
 
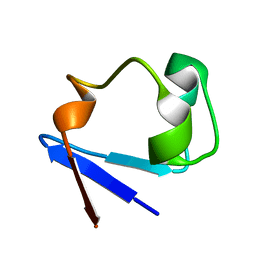 | | 128 hours neutron structure of perdeuterated rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6PH1
 
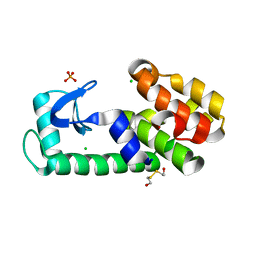 | | T4 lysozyme pseudo-wild type soaked in TEMPOL | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
