3CEZ
 
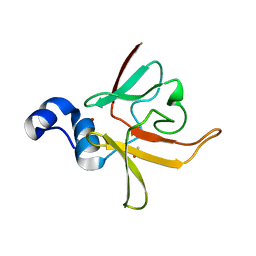 | | Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei | | Descriptor: | ACETIC ACID, Methionine-R-sulfoxide reductase, ZINC ION | | Authors: | Staker, B, Napuli, A, Nakazawa, S.H, Castaneda, L, Alkafeef, S, Vanvoorhis, W, Stewart, L, Myler, P, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine-R-sulfoxide reductase from Burkholderia pseudomallei.
To be Published
|
|
8FBP
 
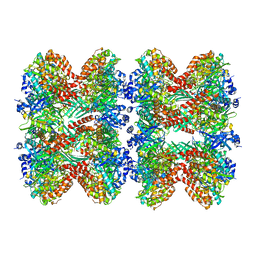 | | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation | | Descriptor: | Glutamine synthetase | | Authors: | Phan, I.Q, Staker, B, Shek, R, Moser, T.H, Evans, J.E, van Voorhis, W.C, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation
To Be Published
|
|
3GMT
 
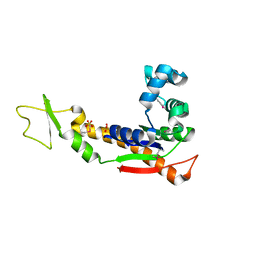 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-03-15 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
8VKA
 
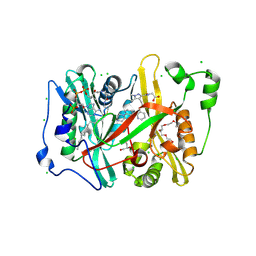 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 9c | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8VKB
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 10b | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8FBQ
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 12b | | Descriptor: | 1-[(3M)-3-{3-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]pyridin-2-yl}phenyl]piperazine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Staker, B.L, Lovell, S.W, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of potent and selective N-myristoyltransferase inhibitors of Plasmodium vivax liver stage hypnozoites and schizonts.
Nat Commun, 14, 2023
|
|
