5CBQ
 
 | |
5BNK
 
 | | Crystal structure of T75C mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-26 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BRB
 
 | | Crystal structure of Q64E mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-30 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BMW
 
 | | Crystal structure of T75V mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BZ4
 
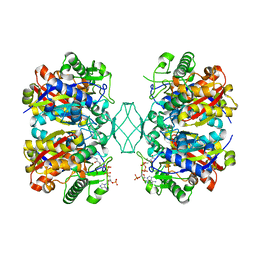 | | Crystal structure of a T1-like thiolase (CoA-complex) from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase, COENZYME A | | Authors: | Janardan, N, Harijan, R.K, Kiema, T.R, Wierenga, R.K, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4G9O
 
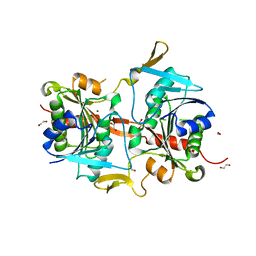 | |
3PVF
 
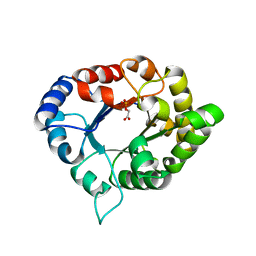 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase complexed with PGA | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
3PY2
 
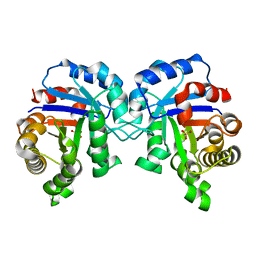 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
4JEX
 
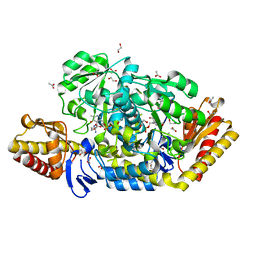 | | Y21K mutant of N-acetylornithine aminotransferase complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4JEZ
 
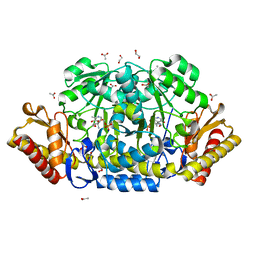 | | N79R mutant of N-acetylornithine aminotransferase complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
3PWA
 
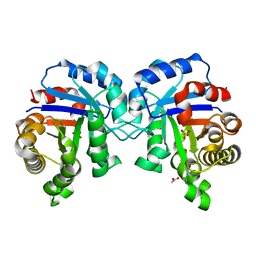 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
4JEV
 
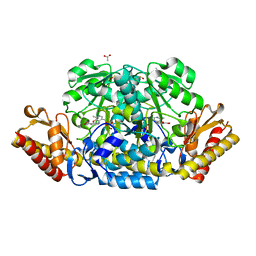 | | N-acetylornithine aminotransferase from S. typhimurium complexed with gabaculine | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases.
To be Published
|
|
4JF1
 
 | | R144Q mutant of N-acetylornithine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4GAD
 
 | |
4JF0
 
 | | N79R mutant of N-acetylornithine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4JEW
 
 | | N-acetylornithine aminotransferase from S. typhimurium complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases.
To be Published
|
|
4JEY
 
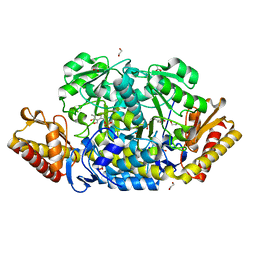 | | E198A mutant of N-acetylornithine aminotransferase from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4Y4Y
 
 | |
4Y5Z
 
 | |
4D9E
 
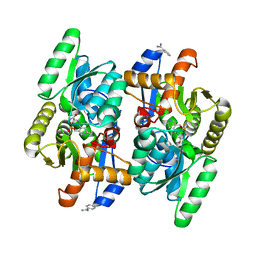 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with L-cycloserine (LCS) | | Descriptor: | BENZAMIDINE, D-Cysteine desulfhydrase, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D96
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with 1-amino-1-carboxycyclopropane (ACC) | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9I
 
 | | Crystal structure of holo Diaminopropionate ammonia lyase from Escherichia coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diaminopropionate ammonia-lyase | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
4D92
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase soaked with beta-chloro-D-alanine shows pyruvate bound 4 A away from active site | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, PYRUVIC ACID, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9K
 
 | | Crystal structure of Escherichia coli Diaminopropionate ammonia lyase in apo form | | Descriptor: | Diaminopropionate ammonia-lyase, PHOSPHATE ION, SULFATE ION | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
4D99
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase with L-ser bound non-covalently at the active site | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, SERINE, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
