4R2M
 
 | |
4RYU
 
 | |
4RYT
 
 | |
2Z6R
 
 | | Crystal structure of Lys49 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N. | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2XPJ
 
 | |
5BYV
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BZ4
 
 | | Crystal structure of a T1-like thiolase (CoA-complex) from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase, COENZYME A | | Authors: | Janardan, N, Harijan, R.K, Kiema, T.R, Wierenga, R.K, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5CBQ
 
 | |
5F0V
 
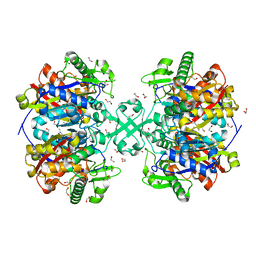 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-11-28 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5F38
 
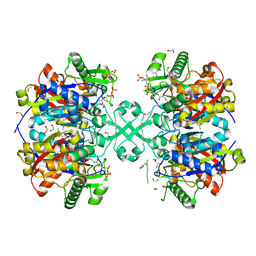 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase, COENZYME A, ... | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-12-02 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5H3L
 
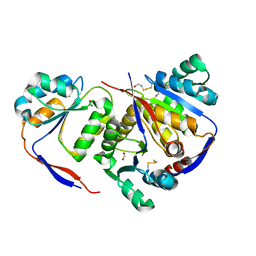 | | Structure of methylglyoxal synthase crystallised as a contaminant | | Descriptor: | FORMIC ACID, Methylglyoxal synthase | | Authors: | Hatti, K, Dadireddy, V, Srinivasan, N, Ramakumar, S, Murthy, M.R.N. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
5H4F
 
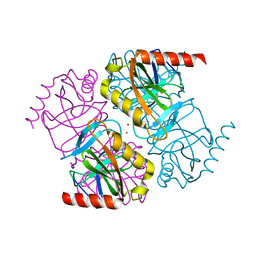 | | Structure of inorganic pyrophosphatase crystallised as a contaminant | | Descriptor: | ZINC ION, inorganic pyrophosphatase | | Authors: | Chaudhary, S, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
5GV4
 
 | |
5H4G
 
 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 1.77 A resolution | | Descriptor: | Ribonuclease VapC4, ZINC ION | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5GZP
 
 | |
5H4H
 
 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 2.2 A resolution | | Descriptor: | CADMIUM ION, Ribonuclease VapC4 | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
4XGB
 
 | | Crystal Structure of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium co-crystallized with AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'/3'-nucleotidase SurE, MAGNESIUM ION, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2014-12-30 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XGP
 
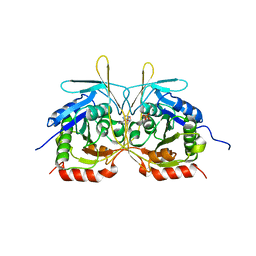 | | Crystal Structure of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium co-crystallized and soaked with AMP. | | Descriptor: | 1,2-ETHANEDIOL, 5'/3'-nucleotidase SurE, ADENINE, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2015-01-01 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XEP
 
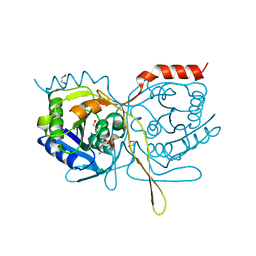 | | Crystal Structure of F222 form of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, 5'/3'-nucleotidase SurE, MAGNESIUM ION, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2014-12-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJ7
 
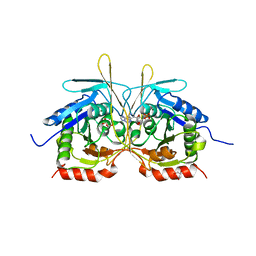 | |
4XER
 
 | | Crystal Structure of C2 form of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'/3'-nucleotidase SurE, ACETATE ION, ... | | Authors: | Mathiharan, Y.K, Murthy, M.R.N. | | Deposit date: | 2014-12-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XH8
 
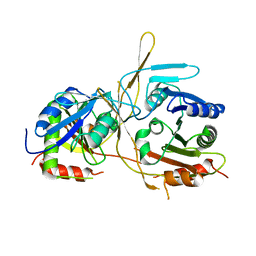 | |
1E57
 
 | |
4X22
 
 | | Crystal structure of Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
2PB2
 
 | | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding | | Descriptor: | 1,2-ETHANEDIOL, Acetylornithine/succinyldiaminopimelate aminotransferase, GLYCEROL, ... | | Authors: | Rajaram, V, Ratna Prasuna, P, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2007-03-28 | | Release date: | 2007-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: Studies on substrate specificity and inhibitor binding
Proteins, 70, 2007
|
|
