5Z6B
 
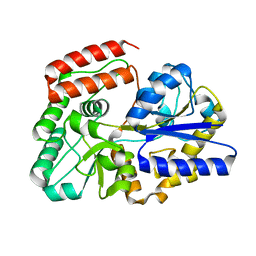 | | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide | | 分子名称: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, Putative ABC transporter substrate-binding protein YesO | | 著者 | Sugiura, H, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2018-01-22 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.582 Å) | | 主引用文献 | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide
To Be Published
|
|
5Z6C
 
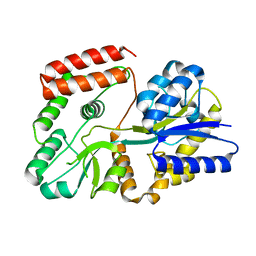 | |
1VAV
 
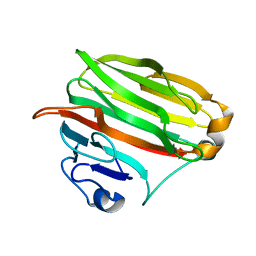 | | Crystal structure of alginate lyase PA1167 from Pseudomonas aeruginosa at 2.0 A resolution | | 分子名称: | Alginate lyase PA1167 | | 著者 | Yamasaki, M, Moriwaki, S, Miyake, O, Hashimoto, W, Murata, K, Mikami, B. | | 登録日 | 2004-02-19 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a beta-sandwich fold.
J.Biol.Chem., 279, 2004
|
|
1VD5
 
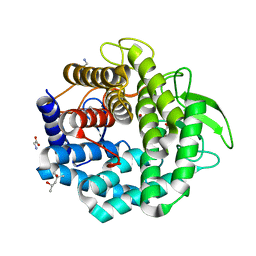 | | Crystal Structure of Unsaturated Glucuronyl Hydrolase, Responsible for the Degradation of Glycosaminoglycan, from Bacillus sp. GL1 at 1.8 A Resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCINE, ... | | 著者 | Itoh, T, Akao, S, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2004-03-18 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Unsaturated Glucuronyl Hydrolase, Responsible for the Degradation of Glycosaminoglycan, from Bacillus sp. GL1 at 1.8 A Resolution
J.Biol.Chem., 279, 2004
|
|
1WOQ
 
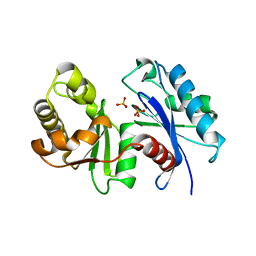 | | Crystal Structure of Inorganic Polyphosphate/ATP-Glucomannokinase From Arthrobacter sp. strain KM At 1.8 A Resolution | | 分子名称: | Inorganic polyphosphate/ATP-glucomannokinase, PHOSPHATE ION, beta-D-glucopyranose | | 著者 | Mukai, T, Kawai, S, Mori, S, Mikami, B, Murata, K. | | 登録日 | 2004-08-24 | | 公開日 | 2004-09-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Bacterial Inorganic Polyphosphate/ATP-glucomannokinase: INSIGHTS INTO KINASE EVOLUTION
J.Biol.Chem., 279, 2004
|
|
1X1J
 
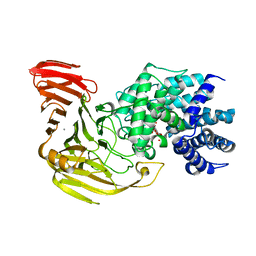 | | Crystal Structure of Xanthan Lyase (N194A) with a Substrate. | | 分子名称: | (4AR,6R,7S,8R,8AR)-8-((5R,6R)-3-CARBOXY-TETRAHYDRO-4,5,6-TRIHYDROXY-2H-PYRAN-2-YLOXY)-HEXAHYDRO-6,7-DIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID), CALCIUM ION, xanthan lyase | | 著者 | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2005-04-04 | | 公開日 | 2005-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
1X1H
 
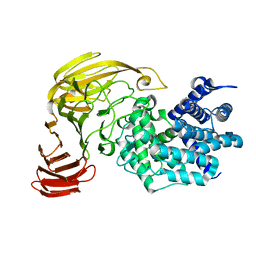 | |
3VR0
 
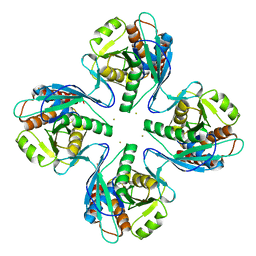 | | Crystal structure of Pyrococcus furiosus PbaB, an archaeal proteasome activator | | 分子名称: | GOLD ION, Putative uncharacterized protein | | 著者 | Kumoi, K, Satoh, T, Hiromoto, T, Mizushima, T, Kamiya, Y, Noda, M, Uchiyama, S, Murata, K, Yagi, H, Kato, K. | | 登録日 | 2012-04-02 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | An archaeal homolog of proteasome assembly factor functions as a proteasome activator
Plos One, 8, 2013
|
|
1X1I
 
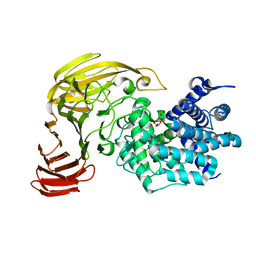 | | Crystal Structure of Xanthan Lyase (N194A) Complexed with a Product | | 分子名称: | (4AR,6R,7S,8R,8AS)-HEXAHYDRO-6,7,8-TRIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID, xanthan lyase | | 著者 | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2005-04-04 | | 公開日 | 2005-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
4Z9X
 
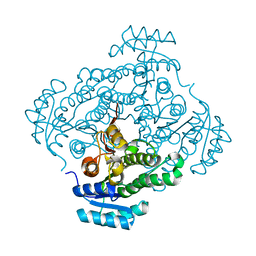 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus pyogenes | | 分子名称: | Gluconate 5-dehydrogenase | | 著者 | Maruyama, Y, Takase, R, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2015-04-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4ZA2
 
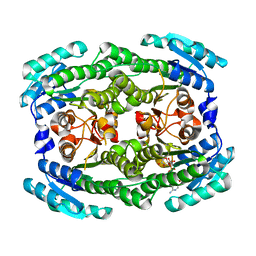 | | Crystal structure of Pectobacterium carotovorum 2-keto-3-deoxy-D-gluconate dehydrogenase complexed with NAD+ | | 分子名称: | 2-deoxy-D-gluconate 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2015-04-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
7DN2
 
 | | Acidic stable capsid structure of Helicobacter pylori bacteriophage KHP30 | | 分子名称: | Cement protein gp15, Major structural protein ORF14 | | 著者 | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2020-12-08 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7F2P
 
 | | The head structure of Helicobacter pylori bacteriophage KHP40 | | 分子名称: | Cement protein gp16, KHP40 MCP | | 著者 | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2021-06-13 | | 公開日 | 2021-10-27 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
2CWS
 
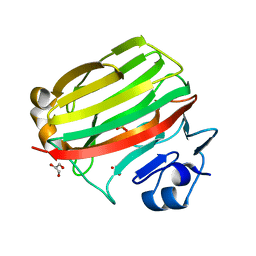 | | Crystal structure at 1.0 A of alginate lyase A1-II', a member of polysaccharide lyase family-7 | | 分子名称: | GLYCEROL, SULFATE ION, alginate lyase A1-II' | | 著者 | Yamasaki, M, Ogura, K, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2005-06-25 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | A Structural Basis for Depolymerization of Alginate by Polysaccharide Lyase Family-7
J.Mol.Biol., 352, 2005
|
|
7DOU
 
 | | Trimeric cement protein structure of Helicobacter pylori bacteriophage KHP40 | | 分子名称: | Cement protein gp16 | | 著者 | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
3WSC
 
 | |
3A1K
 
 | | Crystal structure of Rhodococcus sp. N771 Amidase | | 分子名称: | Amidase | | 著者 | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | 登録日 | 2009-04-09 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition
Biochim.Biophys.Acta, 1804, 2010
|
|
3A1I
 
 | | Crystal structure of Rhodococcus sp. N-771 Amidase complexed with Benzamide | | 分子名称: | Amidase, BENZAMIDE | | 著者 | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | 登録日 | 2009-04-03 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure and Characterization of Amidase from Rhodococcus sp. N-771: Insight into the Molecular Mechanism of Substrate Recognition
Biochim.Biophys.Acta, 2009
|
|
7E55
 
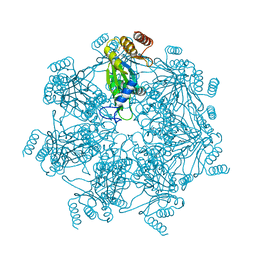 | | Cryo-EM structure of alpha 7 homo-tetradecamer | | 分子名称: | Proteasome subunit alpha type-3 | | 著者 | Song, C, Murata, K. | | 登録日 | 2021-02-17 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Structural Fluctuations of the Human Proteasome alpha 7 Homo-Tetradecamer Double Ring Imply the Proteasomal alpha-Ring Assembly Mechanism.
Int J Mol Sci, 22, 2021
|
|
3VXD
 
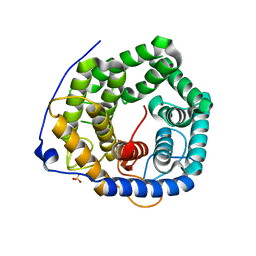 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae | | 分子名称: | Putative uncharacterized protein gbs1889, SULFATE ION | | 著者 | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2012-09-11 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae
To be Published
|
|
3VLW
 
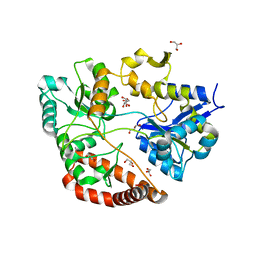 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with mannuronate-guluronate disaccharide | | 分子名称: | AlgQ1, CALCIUM ION, GLYCEROL, ... | | 著者 | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2011-12-05 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
2Z42
 
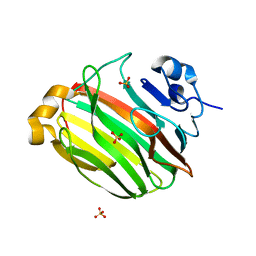 | | Crystal Structure of Family 7 Alginate Lyase A1-II' from Sphingomonas sp. A1 | | 分子名称: | Alginate lyase, SULFATE ION | | 著者 | Ogura, K, Yamasaki, M, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2007-06-12 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Substrate Recognition by Family 7 Alginagte Lyase from Sphingomonas sp. A1
To be published
|
|
2ZAB
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Cmplex with Product (GGG) | | 分子名称: | Alginate lyase, GLYCEROL, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | 著者 | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2007-10-02 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZAC
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Complex with Product (MMG) | | 分子名称: | Alginate lyase, GLYCEROL, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | 著者 | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2007-10-02 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZA9
 
 | | Crystal Structure of Alginate lyase A1-II' N141C/N199C | | 分子名称: | Alginate lyase, SULFATE ION | | 著者 | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2007-10-02 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
