5WKA
 
 | | Crystal structure of a GH1 beta-glucosidase retrieved from microbial metagenome of Poraque Amazon lake | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Morais, M.A.B, Toyama, D, Ramos, F.C, Zanphorlin, L.M, Tonoli, C.C.C, Miranda, F.P, Ruller, R, Henrique-Silva, F, Murakami, M.T. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A novel beta-glucosidase isolated from the microbial metagenome of Lake Poraque (Amazon, Brazil).
Biochim. Biophys. Acta, 1866, 2018
|
|
4W84
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W7U
 
 | |
4W87
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
8VA3
 
 | | Crystal structure of CapGH3b enzyme retrieved from capybara gut metagenome | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Martins, M.P, Morais, M.A.B, Chinaglia, M, Mandelli, F, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
8VA7
 
 | | Crystal structure of CapGH3a enzyme retrieved from capybara gut metagenome | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycoside hydrolase family 3, ... | | Authors: | Martins, M.P, Vieira, P.S, Morais, M.A.B, Mandelli, F, Chinaglia, M, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
8VA4
 
 | | Crystal structure of CapGH16_3 enzyme retrieved from capybara gut metagenome | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 16, PHOSPHATE ION | | Authors: | Vieira, P.S, Martins, M.P, Morais, M.A.B, Mandelli, F, Chinaglia, M, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
6EFU
 
 | | Crystal structure of the double mutant L167W / P172L of the beta-glucosidase from Trichoderma harzianum | | Descriptor: | Beta-glucosidase, NITRATE ION | | Authors: | Morais, M.A.B, Santos, C.A, Tonoli, C.C.C, Souza, A.P, Murakami, M.T. | | Deposit date: | 2018-08-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An engineered GH1 beta-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials.
Sci Rep, 9, 2019
|
|
6EBC
 
 | | OhrB (Organic Hydroperoxide Resistance protein) wild type from Chromobacterium violaceum and reduced by DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6EBG
 
 | | Ohr (Organic Hydroperoxide Resistance protein) mutant - C60S interacting with dihydrolipoamide | | Descriptor: | (6S)-6,8-disulfanyloctanamide, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6EBD
 
 | | OhrB (Organic Hydroperoxide Resistance protein) mutant (C60A) from Chromobacterium violaceum, interacting with dihydrolipoamide | | Descriptor: | (6S)-6,8-disulfanyloctanamide, CHLORIDE ION, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6EB4
 
 | | OhrB (Organic Hydroperoxide Resistance protein) from Chromobacterium violaceum | | Descriptor: | DI(HYDROXYETHYL)ETHER, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-04 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
5JH1
 
 | | Crystal structure of the apo form of AKR4C7 from maize | | Descriptor: | Aldose reductase, AKR4C7 | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5JVO
 
 | | Crystal structure of the Arginine Repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis | | Descriptor: | Arginine repressor, SULFATE ION, TYROSINE | | Authors: | Mariutti, R.B, Ullah, A, Murakami, M.T, Arni, R.K. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine binding and promiscuity in the arginine repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis.
Biochem.Biophys.Res.Commun., 475, 2016
|
|
5KB6
 
 | | High-resolution structure of the adenosine kinase from Mus musculus in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, Adenosine kinase, ... | | Authors: | Oliveira, R.R, Neto, R.M, Polo, C.C, Tonoli, C.C.C, Murakami, M.T, Franchini, K.G. | | Deposit date: | 2016-06-02 | | Release date: | 2017-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of the adenosine kinase from Mus musculus in complex with adenosine
To Be Published
|
|
5KB5
 
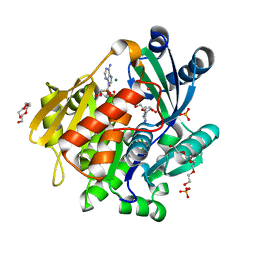 | | Crystal structure of the adenosine kinase from Mus musculus in complex with adenosine and adenosine-diphosphate | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Adenosine kinase, ... | | Authors: | Oliveira, R.R, Neto, R.M, Polo, C.C, Tonoli, C.C.C, Murakami, M.T, Franchini, K.G. | | Deposit date: | 2016-06-02 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the adenosine kinase from Mus musculus in complex with adenosine and adenosine-diphosphate
To Be Published
|
|
5JGW
 
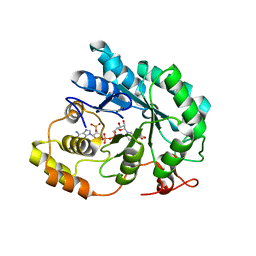 | | Crystal structure of maize AKR4C13 in complex with NADP and acetate | | Descriptor: | ACETATE ION, Aldose reductase, AKR4C13, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of maize AKR4C13 in complex with NADP and acetate
To Be Published
|
|
3GBO
 
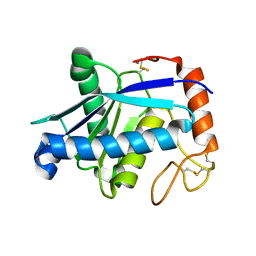 | | Crystal structure of BmooMPalpha-I, a non-hemorrhagic metalloproteinase isolated from Bothrops moojeni snake venom | | Descriptor: | CALCIUM ION, ZINC ION, Zinc metalloproteinase BmooMPalfa-I | | Authors: | Akao, P.K, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2009-02-20 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural studies of BmooMPalpha-I, a non-hemorrhagic metalloproteinase from Bothrops moojeni venom.
Toxicon, 55, 2010
|
|
5HPC
 
 | |
5HNN
 
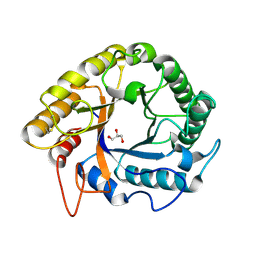 | |
5HOS
 
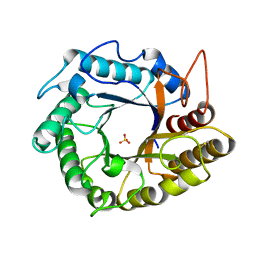 | |
2L8A
 
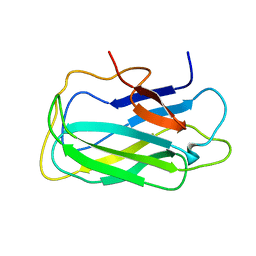 | | Structure of a novel CBM3 lacking the calcium-binding site | | Descriptor: | Endoglucanase | | Authors: | Paiva, J.H, Meza, A.N, Sforca, M.L, Navarro, R.Z, Neves, J.L, Santos, C.R, Murakami, M.T, Zeri, A.C. | | Deposit date: | 2011-01-07 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
2KQ5
 
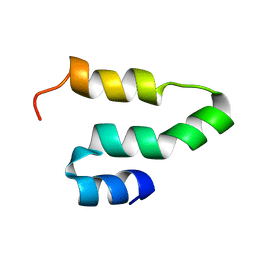 | |
2P3F
 
 | | Crystal structure of the factor Xa/NAP5 complex | | Descriptor: | Anti-coagulant protein 5, Coagulation factor X, SODIUM ION | | Authors: | Rios-Steiner, J.L, Murakami, M.T, Tulinsky, A, Arni, R.K. | | Deposit date: | 2007-03-08 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Active and exo-site inhibition of human factor Xa: structure of des-Gla factor Xa inhibited by NAP5, a potent nematode anticoagulant protein from Ancylostoma caninum
J.Mol.Biol., 371, 2007
|
|
4RW3
 
 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | D-MYO-INOSITOL-1-PHOSPHATE, DECANOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
