6XKU
 
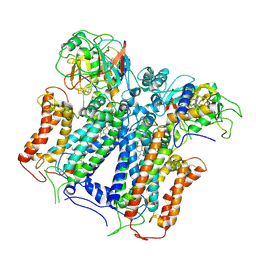 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKZ
 
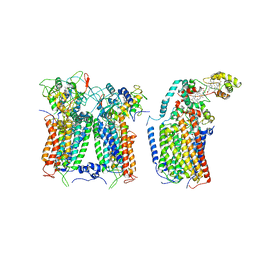 | | R. capsulatus CIII2CIV tripartite super-complex, conformation B (SC-1B) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKW
 
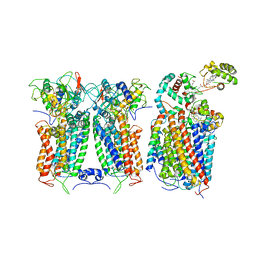 | | R. capsulatus CIII2CIV bipartite super-complex (SC-2A) with CcoH/cy | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKX
 
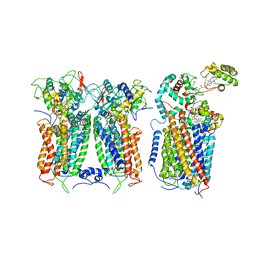 | | R. capsulatus CIII2CIV tripartite super-complex, conformation A (SC-1A) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6DT7
 
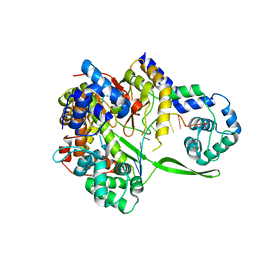 | | Bacteriophage N4 RNA polymerase II and DNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*CP*TP*GP*CP*A)-3'), RNAP1, RNAP2 | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
6DT8
 
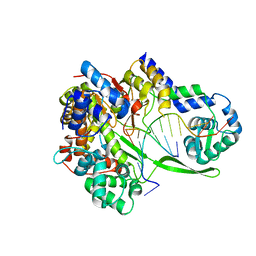 | | Bacteriophage N4 RNA polymerase II elongation complex 1 | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), RNA (5'-R(P*UP*GP*GP*UP*GP*G)-3'), RNAP1, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
6DTA
 
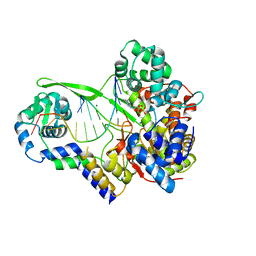 | | Bacteriophage N4 RNA polymerase II elongation complex 2 | | Descriptor: | DNA (5'-D(P*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
3G37
 
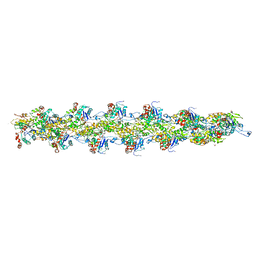 | | Cryo-EM structure of actin filament in the presence of phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wakabayshi, T, Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q. | | Deposit date: | 2009-02-02 | | Release date: | 2010-11-03 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
8SYI
 
 | | Cyanobacterial RNAP-EC | | Descriptor: | DNA (37-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Qayyum, M.Z, Imashimizu, M, Leanca, M, Vishwakarma, R.K, Bradley Riaz, A, Yuzenkova, Y, Murakami, K.S. | | Deposit date: | 2023-05-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8URW
 
 | | Cyanobacterial RNA polymerase elongation complex with NusG and CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (40-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Imashimizu, M, Leanca, M, Vishwakarma, R.K, Bradley Riaz, A, Yuzenkova, Y, Murakami, K.S. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7R8J
 
 | |
7R8H
 
 | |
5EZK
 
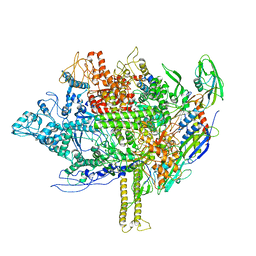 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
7R8F
 
 | |
7R8K
 
 | |
7R8G
 
 | |
7R8I
 
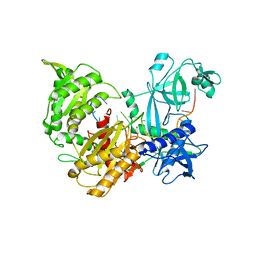 | |
6KF9
 
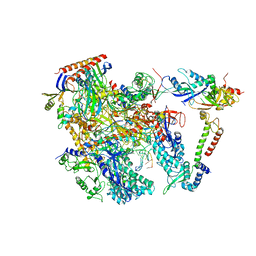 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF4
 
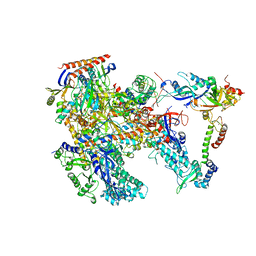 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF3
 
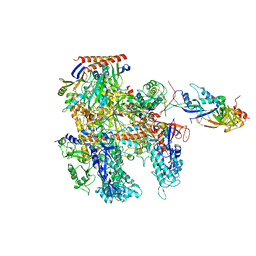 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
8GHA
 
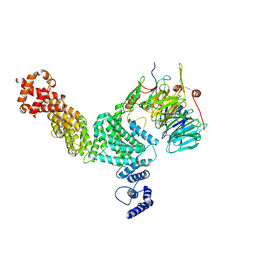 | | Hir3 Arm/Tail, Hir2 WD40, C-terminal Hpc2 | | Descriptor: | Histone promoter control protein 2, Histone transcription regulator 3, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
8GHM
 
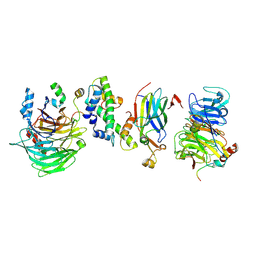 | | Hir1 WD40 domains and Asf1/H3/H4 | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
6OY7
 
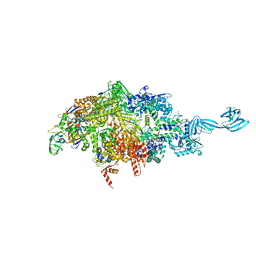 | |
6OVY
 
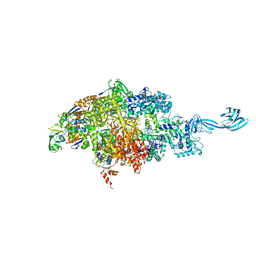 | |
6OVR
 
 | |
