6WTD
 
 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc with inhibitor of Bedaquiline bound | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Mueller, D.M, Srivastava, A.P, Symersky, J, Luo, M, Liao, M.F. | | Deposit date: | 2020-05-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Bedaquiline inhibits the yeast and human mitochondrial ATP synthases.
Commun Biol, 3, 2020
|
|
8FKJ
 
 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FL8
 
 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F29
 
 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F39
 
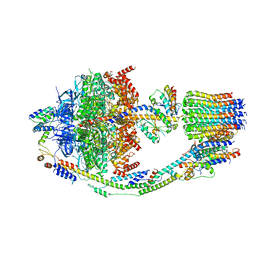 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3OE7
 
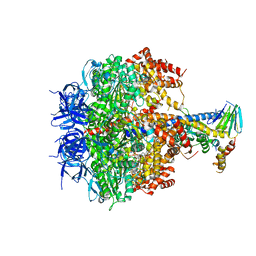 | | Structure of four mutant forms of yeast f1 ATPase: gamma-I270T | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3OFN
 
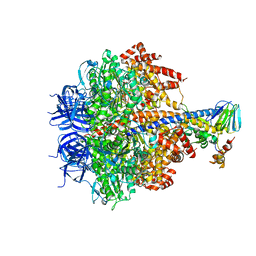 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3OEE
 
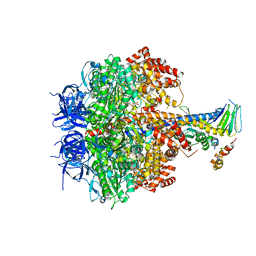 | | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S
To be Published
|
|
5BQ6
 
 | |
5BQA
 
 | |
5BQJ
 
 | |
5BPS
 
 | |
6CP6
 
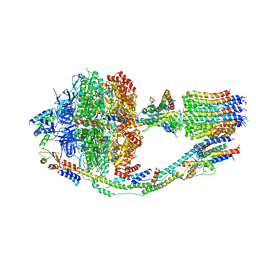 | | Monomer yeast ATP synthase (F1Fo) reconstituted in nanodisc. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
6CP7
 
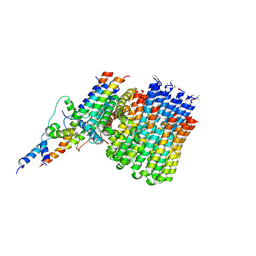 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement. | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
6CP5
 
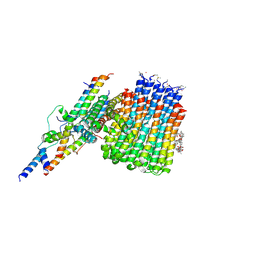 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc with inhibitor of oligomycin bound generated from focused refinement. | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
6CP3
 
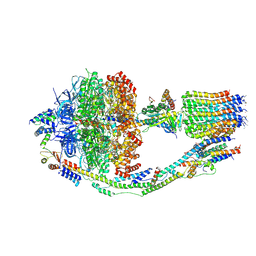 | | Monomer yeast ATP synthase (F1Fo) reconstituted in nanodisc with inhibitor of oligomycin bound. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
3OEH
 
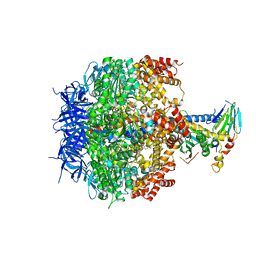 | | Structure of four mutant forms of yeast F1 ATPase: beta-V279F | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
2HLD
 
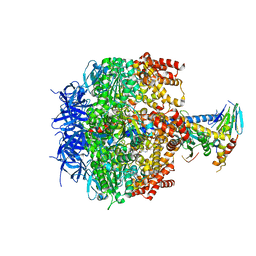 | | Crystal structure of yeast mitochondrial F1-ATPase | | Descriptor: | ATP synthase alpha chain, mitochondrial, ATP synthase beta chain, ... | | Authors: | Kabaleeswaran, V, Puri, N, Walker, J.E, Leslie, A.G, Mueller, D.M. | | Deposit date: | 2006-07-06 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel features of the rotary catalytic mechanism revealed in the structure of yeast F(1) ATPase.
Embo J., 25, 2006
|
|
3U2Y
 
 | | ATP synthase c10 ring in proton-unlocked conformation at pH 6.1 | | Descriptor: | ATP synthase subunit C, mitochondrial | | Authors: | Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, J, Mueller, D.M. | | Deposit date: | 2011-10-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3U32
 
 | | ATP synthase c10 ring reacted with DCCD at pH 5.5 | | Descriptor: | ATP synthase subunit C, mitochondrial, DICYCLOHEXYLUREA | | Authors: | Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, J, Mueller, D.M. | | Deposit date: | 2011-10-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3U2F
 
 | | ATP synthase c10 ring in proton-unlocked conformation at PH 8.3 | | Descriptor: | ATP synthase subunit C, mitochondrial | | Authors: | Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, J, Mueller, D.M. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3FKS
 
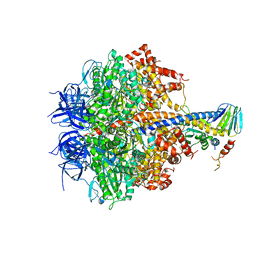 | | Yeast F1 ATPase in the absence of bound nucleotides | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Kabaleeswaran, V, Symersky, J, Shen, H, Walker, J.E, Leslie, A.G.W, Mueller, D.M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.587 Å) | | Cite: | Asymmetric structure of the yeast f1 ATPase in the absence of bound nucleotides.
J.Biol.Chem., 284, 2009
|
|
4F4S
 
 | |
3ZIA
 
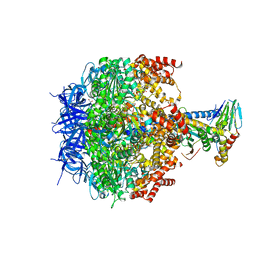 | | The structure of F1-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Robinson, G.C, Bason, J.V, Montgomery, M.G, Fearnley, I.M, Mueller, D.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2013-01-07 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of F1-ATPase from Saccharomyces Cerevisiae Inhibited by its Regulatory Protein If1.
Open Biol., 3, 2013
|
|
