1PGR
 
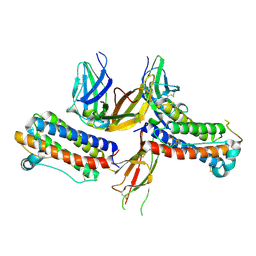 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
4XSL
 
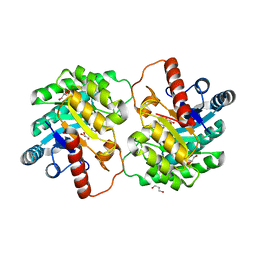 | | Crystal strcutre of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with glycerol | | Descriptor: | D-tagatose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
2DSO
 
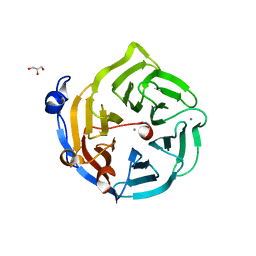 | | Crystal structure of D138N mutant of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Drp35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-07-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DG0
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | DrP35 | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
4Q0V
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
2N9I
 
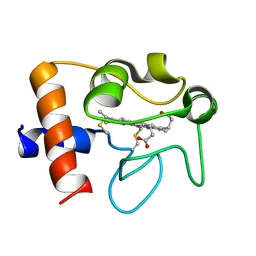 | | Solution structure of reduced human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2N9J
 
 | | Solution structure of oxidized human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1VAS
 
 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | Authors: | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
4XSM
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | Descriptor: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5H6O
 
 | | Porphobilinogen deaminase from Vibrio Cholerae | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, MAGNESIUM ION, Porphobilinogen deaminase | | Authors: | Funamizu, T, Chen, M, Tanaka, Y, Ishimori, K, Uchida, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Porphobilinogen deaminase from Vibrio Cholerae
To Be Published
|
|
5E1U
 
 | | Crystal structure of IrCp*-apo-Fr | | Descriptor: | CADMIUM ION, Ferritin light chain, IRIDIUM (III) ION, ... | | Authors: | Maity, B, Fukumori, K, Abe, S, Ueno, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Immobilization of two organometallic complexes into a single cage to construct protein-based microcompartments
Chem.Commun.(Camb.), 52, 2016
|
|
4Q0Q
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
5E2D
 
 | | Crystal structure of IrCp*/Pd(allyl)-apo-Fr | | Descriptor: | CADMIUM ION, Ferritin light chain, IRIDIUM (III) ION, ... | | Authors: | Maity, B, Fukumori, K, Abe, S, Ueno, T. | | Deposit date: | 2015-10-01 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Immobilization of two organometallic complexes into a single cage to construct protein-based microcompartments
Chem.Commun.(Camb.), 52, 2016
|
|
2DG1
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus, complexed with Ca2+ | | Descriptor: | CALCIUM ION, DrP35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
1EQ5
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQE
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-04 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
4Q0S
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with ribitol | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), D-ribitol, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0P
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
1GOB
 
 | | COOPERATIVE STABILIZATION OF ESCHERICHIA COLI RIBONUCLEASE HI BY INSERTION OF GLY-80B AND GLY-77-> ALA SUBSTITUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Nakamura, H, Morikawa, K, Kanaya, S. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77-->Ala substitution.
Biochemistry, 32, 1993
|
|
1GOC
 
 | | COOPERATIVE STABILIZATION OF ESCHERICHIA COLI RIBONUCLEASE HI BY INSERTION OF GLY-80B AND GLY-77-> ALA SUBSTITUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Nakamura, H, Morikawa, K, Kanaya, S. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77-->Ala substitution.
Biochemistry, 32, 1993
|
|
1GOA
 
 | | COOPERATIVE STABILIZATION OF ESCHERICHIA COLI RIBONUCLEASE HI BY INSERTION OF GLY-80B AND GLY-77-> ALA SUBSTITUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Nakamura, H, Morikawa, K, Kanaya, S. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77-->Ala substitution.
Biochemistry, 32, 1993
|
|
4Z2M
 
 | | Crystal structure of human SPT16 Mid-AID/H3-H4 tetramer FACT Histone complex | | Descriptor: | FACT complex subunit SPT16, Histone H3.1, Histone H4 | | Authors: | Tsunaka, Y, Fujiwara, Y, Oyama, T, Hirose, S, Morikawa, K. | | Deposit date: | 2015-03-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Integrated molecular mechanism directing nucleosome reorganization by human FACT.
Genes Dev., 30, 2016
|
|
1FUT
 
 | | CRYSTAL STRUCTURES OF RIBONUCLEASE F1 OF FUSARIUM MONILIFORME IN ITS FREE FORM AND IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE F1 | | Authors: | Katayanagi, K, Vassylyev, D.G, Ishikawa, K, Morikawa, K. | | Deposit date: | 1993-01-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of ribonuclease F1 of Fusarium moniliforme in its free form and in complex with 2'GMP.
J.Mol.Biol., 230, 1993
|
|
