8J3R
 
 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
5GQI
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Ala194 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQJ
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Ser193 and Ala194 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQK
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQL
 
 | | Crystal structure of Wild Type Cypovirus Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQM
 
 | | Crystal structure of in cellulo Wild Type Cypovirus Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
2OH5
 
 | | The Crystal Structure of Infectious Cypovirus Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Coulibaly, F, Chiu, E, Ikeda, K, Gutmann, S, Haebel, P.W, Schulze-Briese, C, Mori, H, Metcalf, P. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The molecular organization of cypovirus polyhedra.
Nature, 446, 2007
|
|
5GQN
 
 | | Crystal structure of in cellulo Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
8JHH
 
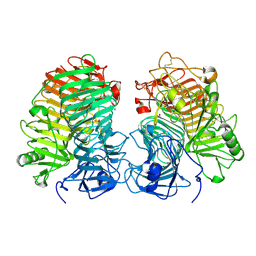 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
2RKB
 
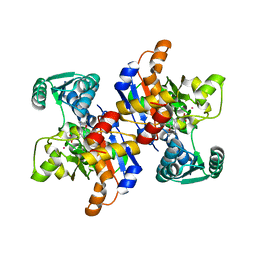 | | Serine dehydratase like-1 from human cancer cells | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine dehydratase-like | | Authors: | Yamada, T, Komoto, J, Kasuya, T, Mori, H, Ogawa, H, Takusagawa, F. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A catalytic mechanism that explains a low catalytic activity of serine dehydratase like-1 from human cancer cells: Crystal structure and site-directed mutagenesis studies.
Biochim.Biophys.Acta, 1780, 2008
|
|
2OH7
 
 | | The Crystal Structure of Cypovirus Polyhedra containing the Human ZIP-kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Coulibaly, F, Chiu, E, Ikeda, K, Gutmann, S, Haebel, P.W, Schulze-Briese, C, Mori, H, Metcalf, P. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The molecular organization of cypovirus polyhedra.
Nature, 446, 2007
|
|
2OH6
 
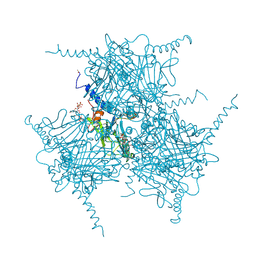 | | The Crystal Structure of Recombinant Cypovirus Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Coulibaly, F, Chiu, E, Ikeda, K, Gutmann, S, Haebel, P.W, Schulze-Briese, C, Mori, H, Metcalf, P. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular organization of cypovirus polyhedra.
Nature, 446, 2007
|
|
5GK2
 
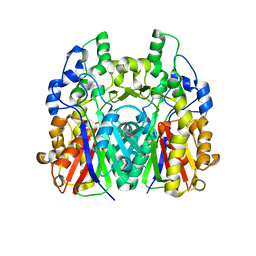 | |
5GK0
 
 | |
5GK1
 
 | | Crystal structure of the ketosynthase StlD complexed with substrate | | Descriptor: | 3-OXO-5-METHYLHEXANOIC ACID, Ketosynthase StlD | | Authors: | Mori, T, Saito, Y, Morita, H, Abe, I. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insight into the Enzymatic Formation of Bacterial Stilbene.
Cell Chem Biol, 23, 2016
|
|
5B0D
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y27W mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0E
 
 | | Polyketide cyclase OAC from Cannabis sativa, V59M mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0F
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y72F mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B09
 
 | | Polyketide cyclase OAC from Cannabis sativa bound with Olivetolic acid | | Descriptor: | 2,4-bis(oxidanyl)-6-pentyl-benzoic acid, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0G
 
 | | Polyketide cyclase OAC from Cannabis sativa, H78S mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0A
 
 | | Polyketide cyclase OAC from Cannabis sativa, H5Q mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B08
 
 | | Polyketide cyclase OAC from Cannabis sativa | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.325 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0C
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y27F mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0B
 
 | | Polyketide cyclase OAC from Cannabis sativa, I7F mutant | | Descriptor: | ACETATE ION, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
