8GQA
 
 | |
1UF8
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-Phenylalanine | | Descriptor: | D-[(AMINO)CARBONYL]PHENYLALANINE, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF4
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase | | Descriptor: | N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid
To be published
|
|
1UF7
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-valine | | Descriptor: | 3-METHYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF5
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1C9Y
 
 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CATALYTIC MECHANISM | | Descriptor: | NORVALINE, ORNITHINE CARBAMOYLTRANSFERASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Yu, X, Morizono, H, Tuchman, M, Allewell, N.M. | | Deposit date: | 1999-08-03 | | Release date: | 2000-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
Proteins, 39, 2000
|
|
1CG5
 
 | | DEOXY FORM HEMOGLOBIN FROM DASYATIS AKAJEI | | Descriptor: | PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chong, K.T, Morimoto, H. | | Deposit date: | 1999-03-26 | | Release date: | 1999-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the deoxy and CO forms of haemoglobin from Dasyatis akajei, a cartilaginous fish.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CG8
 
 | | CO Form Hemoglobin from Dasyatis Akajei | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chong, K.T, Morimoto, H. | | Deposit date: | 1999-03-26 | | Release date: | 1999-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the deoxy and CO forms of haemoglobin from Dasyatis akajei, a cartilaginous fish.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1UL1
 
 | | Crystal structure of the human FEN1-PCNA complex | | Descriptor: | Flap endonuclease-1, MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Sakurai, S, Kitano, K, Yamaguchi, H, Hamada, K, Okada, K, Fukuda, K, Uchida, M, Ohtsuka, E, Morioka, H, Hakoshima, T. | | Deposit date: | 2003-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for recruitment of human flap endonuclease 1 to PCNA
EMBO J., 24, 2005
|
|
2R8V
 
 | | Native structure of N-acetylglutamate synthase from Neisseria gonorrhoeae | | Descriptor: | ACETYL COENZYME *A, Putative acetylglutamate synthase | | Authors: | Shi, D, Sagar, V, Jin, Z, Yu, X, Caldovic, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2007-09-11 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
J.Biol.Chem., 283, 2008
|
|
2R98
 
 | | Crystal Structure of N-acetylglutamate synthase (selenoMet substituted) from Neisseria gonorrhoeae | | Descriptor: | ACETYL COENZYME *A, Putative acetylglutamate synthase | | Authors: | Shi, D, Sagar, V, Jin, Z, Yu, X, Caldovic, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
J.Biol.Chem., 283, 2008
|
|
2EWI
 
 | |
2EWU
 
 | |
5AUR
 
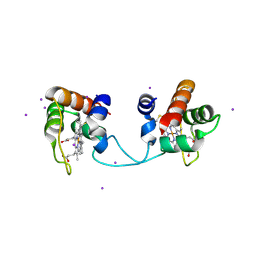 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at N-terminal region | | Descriptor: | Cytochrome c-552, HEME C, IODIDE ION | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Kamikubo, H, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
4OOQ
 
 | | apo-dUTPase from Arabidopsis thaliana | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, SULFATE ION | | Authors: | Inoguchi, N, Bajaj, M, Moriyama, H. | | Deposit date: | 2014-02-03 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
6LFW
 
 | |
6LCS
 
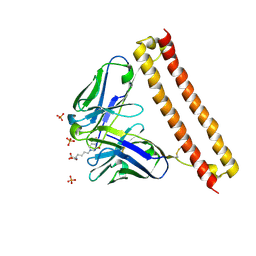 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | Descriptor: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | Authors: | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
6LFV
 
 | |
1OTH
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH N-PHOSPHONACETYL-L-ORNITHINE | | Descriptor: | N-(PHOSPHONOACETYL)-L-ORNITHINE, PROTEIN (ORNITHINE TRANSCARBAMOYLASE) | | Authors: | Shi, D, Morizono, H, Ha, Y, Aoyagi, M, Tuchman, N, Allewell, N.M. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85-A resolution crystal structure of human ornithine transcarbamoylase complexed with N-phosphonacetyl-L-ornithine. Catalytic mechanism and correlation with inherited deficiency.
J.Biol.Chem., 273, 1998
|
|
1OSJ
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1OSI
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
5F9K
 
 | | Dictyostelium discoideum dUTPase at 2.2 Angstrom | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, ... | | Authors: | Inoguchi, N, Chia, C.P, Heck, K, Moriyama, H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Mitochondrial localization of Dictyostelium discoideum dUTPase mediated by its N-terminus.
Bmc Res Notes, 13, 2020
|
|
1XAC
 
 | |
1XAD
 
 | |
2FMY
 
 | |
