3WEM
 
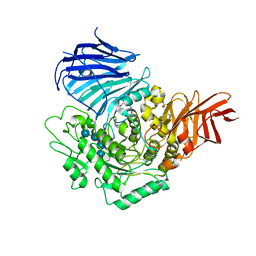 | | Sugar beet alpha-glucosidase with acarviosyl-maltotetraose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, ... | | 著者 | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | 登録日 | 2013-07-09 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.591 Å) | | 主引用文献 | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3W7X
 
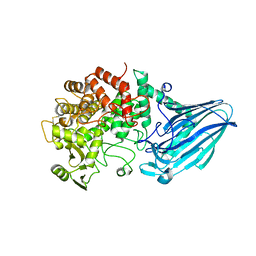 | | Crystal structure of E. coli YgjK D324N complexed with melibiose | | 分子名称: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose | | 著者 | Miyazaki, T, Ichikawa, M, Yokoi, G, Kitaoka, M, Mori, H, Kitano, Y, Nishikawa, A, Tonozuka, T. | | 登録日 | 2013-03-08 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of a bacterial glycoside hydrolase family 63 enzyme in complex with its glycosynthase product, and insights into the substrate specificity.
Febs J., 280, 2013
|
|
3WEL
 
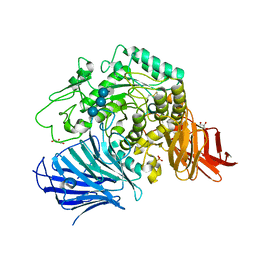 | | Sugar beet alpha-glucosidase with acarviosyl-maltotriose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, GLYCEROL, ... | | 著者 | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | 登録日 | 2013-07-08 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3WEN
 
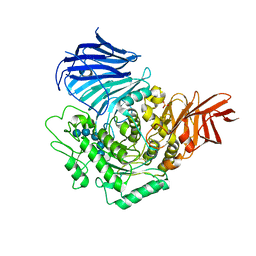 | | Sugar beet alpha-glucosidase with acarviosyl-maltopentaose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, ... | | 著者 | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | 登録日 | 2013-07-09 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3CA9
 
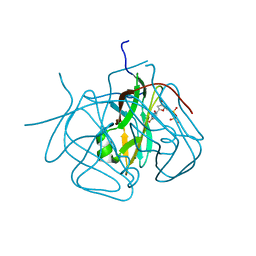 | | Evolution of chlorella virus dUTPase | | 分子名称: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | 著者 | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | 登録日 | 2008-02-19 | | 公開日 | 2009-03-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1VDZ
 
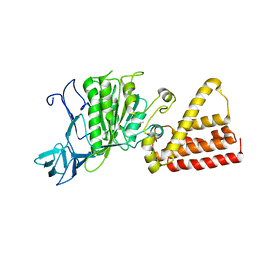 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | 著者 | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | 登録日 | 2004-03-26 | | 公開日 | 2005-06-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
4OOP
 
 | | Arabidopsis thaliana dUTPase with with magnesium and alpha,beta-imido-dUTP | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | 著者 | Inoguchi, N, Bajaj, M, Moriyama, H. | | 登録日 | 2014-02-03 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
1WNI
 
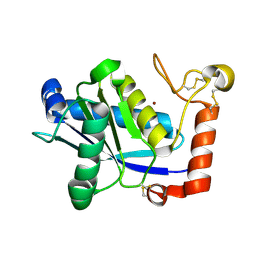 | | Crystal Structure of H2-Proteinase | | 分子名称: | Trimerelysin II, ZINC ION | | 著者 | Kumasaka, T, Yamamoto, M, Moriyama, H, Tanaka, N, Sato, M, Katsube, Y, Yamakawa, Y, Omori-Satoh, T, Iwanaga, S, Ueki, T. | | 登録日 | 2004-08-04 | | 公開日 | 2004-08-17 | | 最終更新日 | 2019-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
J.Biochem., 119, 1996
|
|
6LFX
 
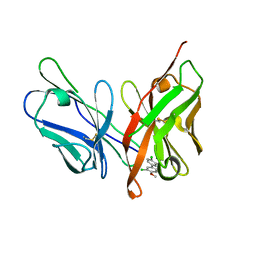 | |
7CIO
 
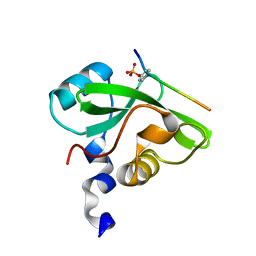 | | Molecular interactions of cytoplasmic region of CTLA-4 with SH2 domains of PI3-kinase | | 分子名称: | Cytotoxic T-lymphocyte protein 4, Phosphatidylinositol 3-kinase regulatory subunit alpha | | 著者 | Iiyama, M, Numoto, N, Ogawa, S, Kuroda, M, Morii, H, Abe, R, Ito, N, Oda, M. | | 登録日 | 2020-07-08 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Molecular interactions of the CTLA-4 cytoplasmic region with the phosphoinositide 3-kinase SH2 domains.
Mol.Immunol., 131, 2021
|
|
2FFN
 
 | |
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | 分子名称: | RIBOSOME RECYCLING FACTOR | | 著者 | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | 登録日 | 2000-10-19 | | 公開日 | 2001-05-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
1QSH
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | 分子名称: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | 登録日 | 1999-06-22 | | 公開日 | 1999-07-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
1QSI
 
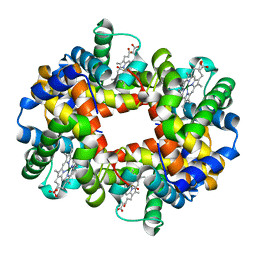 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | 分子名称: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), ... | | 著者 | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | 登録日 | 1999-06-22 | | 公開日 | 1999-07-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
2EWK
 
 | |
3NBS
 
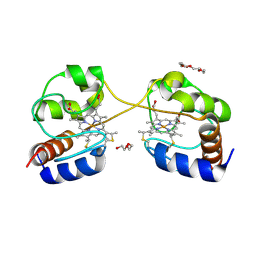 | | Crystal structure of dimeric cytochrome c from horse heart | | 分子名称: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | 著者 | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | 登録日 | 2010-06-04 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NBT
 
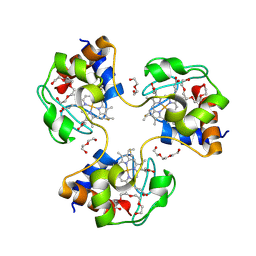 | | Crystal structure of trimeric cytochrome c from horse heart | | 分子名称: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | 著者 | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | 登録日 | 2010-06-04 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1WUJ
 
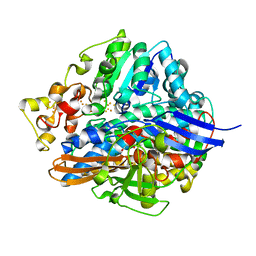 | | Three-Dimensional Structure Of The Ni-B State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | 分子名称: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | 著者 | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | 登録日 | 2004-12-07 | | 公開日 | 2005-12-07 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUH
 
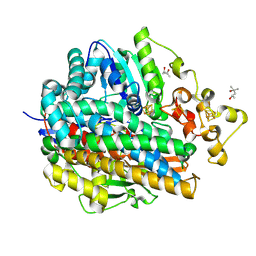 | | Three-Dimensional Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | 登録日 | 2004-12-07 | | 公開日 | 2005-12-07 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Structural basis for the activation process of [NiFe] hydrogenase from D.vulgaris Miyazaki F
To be Published
|
|
5KER
 
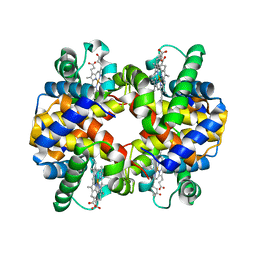 | | Deer mouse recombinant hemoglobin from high altitude species | | 分子名称: | Alpha-globin, Beta globin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Inoguchi, N, Natarajan, C, Storz, J.F, Moriyama, H. | | 登録日 | 2016-06-10 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Alteration of the alpha 1 beta 2/ alpha 2 beta 1 subunit interface contributes to the increased hemoglobin-oxygen affinity of high-altitude deer mice.
PLoS ONE, 12, 2017
|
|
1WUK
 
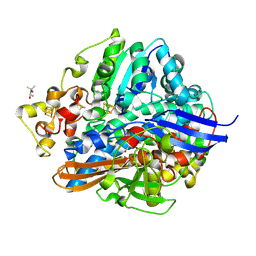 | | High resolution Structure Of The Oxidized State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | 登録日 | 2004-12-07 | | 公開日 | 2005-12-07 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUL
 
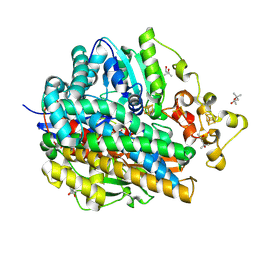 | | High Resolution Structure Of The Reduced State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | 著者 | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | 登録日 | 2004-12-07 | | 公開日 | 2005-12-07 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUI
 
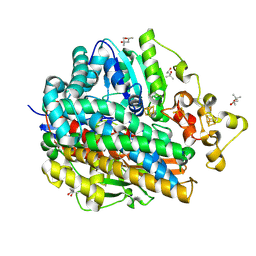 | | Ultra-High resolution Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | 著者 | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | 登録日 | 2004-12-07 | | 公開日 | 2005-12-07 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
8GQ9
 
 | |
8GQB
 
 | |
