3IWK
 
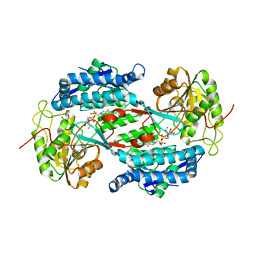 | | Crystal structure of aminoaldehyde dehydrogenase 1 from Pisum sativum (PsAMADH1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Aminoaldehyde dehydrogenase, GLYCEROL, ... | | Authors: | Kopecny, D, Morera, S, Briozzo, P. | | Deposit date: | 2009-09-02 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes.
J.Mol.Biol., 396, 2010
|
|
3S1C
 
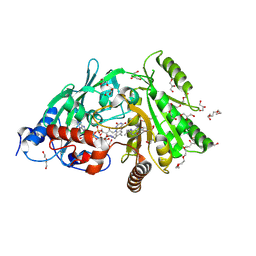 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3S1D
 
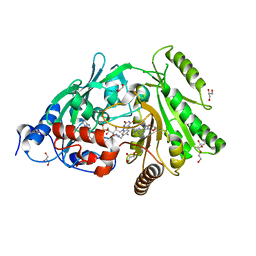 | | Glu381Ser mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3S1F
 
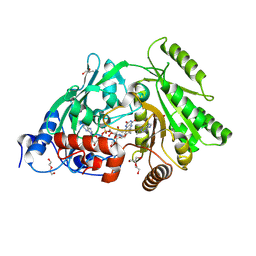 | | Asp169Glu mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3S1E
 
 | | Pro427Gln mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
1KDN
 
 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cherfils, J, Xu, Y.W, Morera, S, Janin, J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5ORE
 
 | |
5OTC
 
 | |
5OT9
 
 | |
5ORG
 
 | |
5OT8
 
 | |
5OTA
 
 | |
4O95
 
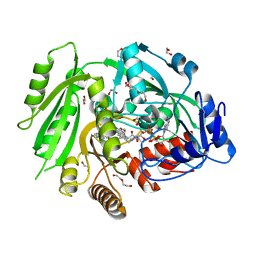 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-01 | | Release date: | 2015-04-01 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
4OAL
 
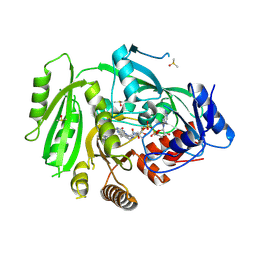 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU in alternative spacegroup | | Descriptor: | 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, DIMETHYL SULFOXIDE, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
1KKM
 
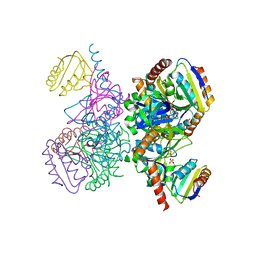 | | L.casei HprK/P in complex with B.subtilis P-Ser-HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHATE ION, ... | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JB1
 
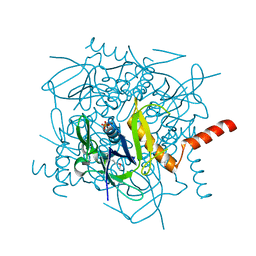 | | Lactobacillus casei HprK/P Bound to Phosphate | | Descriptor: | HPRK PROTEIN, PHOSPHATE ION | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Monedero, V, Gueguen-Chaignon, V, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain.
EMBO J., 20, 2001
|
|
1KKL
 
 | | L.casei HprK/P in complex with B.subtilis HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHOCARRIER PROTEIN HPR | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K0C
 
 | | Ure2p in complex with S-p-nitrobenzylglutathione | | Descriptor: | GLUTATHIONE, S-(P-NITROBENZYL)GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1K0A
 
 | | Ure2p in Complex with S-hexylglutathione | | Descriptor: | GLUTATHIONE, S-HEXYLGLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1K0D
 
 | | Ure2p in Complex with Glutathione | | Descriptor: | GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1K44
 
 | |
1JZR
 
 | | Ure2p in complex with glutathione | | Descriptor: | GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-17 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1K0B
 
 | | Ure2p in Complex with Glutathione | | Descriptor: | GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1M5R
 
 | | Ternary complex of T4 phage BGT with UDP and a 13 mer DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2002-07-10 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Base-flipping mechanism for the T4 phage beta-glucosyltransferase and
identification of a transition state analog
J.Mol.Biol., 324, 2002
|
|
1NDK
 
 | | X-RAY STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Dumas, C, Morera, S, Lascu, I, Veron, M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of nucleoside diphosphate kinase.
EMBO J., 11, 1992
|
|
