5CFE
 
 | | Bacillus subtilis AP endonuclease ExoA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.
Biochimie, 128-129, 2016
|
|
5CFG
 
 | | C2 crystal form of APE1 with Mg2+ | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.
Biochimie, 128-129, 2016
|
|
7R3P
 
 | |
7R4Z
 
 | |
7R5E
 
 | |
7R5P
 
 | |
7R4V
 
 | |
7R3S
 
 | | FtrA/P19 of Rubrivivax gelatinosus in complex with Ni | | Descriptor: | FtrA-P19 protein, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2022-02-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | New insights into the mechanism of iron transport through the bacterial Ftr system present in pathogens.
Febs J., 289, 2022
|
|
7R5G
 
 | |
6QAK
 
 | |
6QAP
 
 | |
6QAO
 
 | |
4PXL
 
 | | Structure of Zm ALDH2-3 (RF2C) in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytosolic aldehyde dehydrogenase RF2C, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-18 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7.
Biochem.J., 468, 2015
|
|
4PXN
 
 | | Structure of Zm ALDH7 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Uncharacterized protein | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7.
Biochem.J., 468, 2015
|
|
8CKD
 
 | | PBP AccA from A. fabrum C58 in complex with agrocinopine D-like | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-15 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C6W
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 1 | | Descriptor: | Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CHC
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine D-like | | Descriptor: | Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, Deicsics, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CDO
 
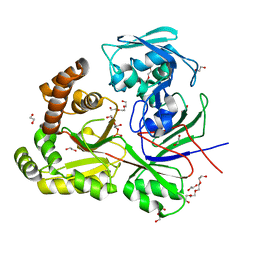 | |
8CI6
 
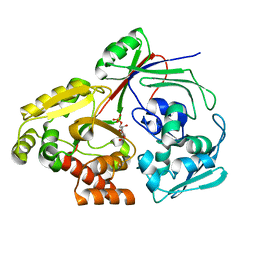 | | PBP AccA from A. vitis S4 in complex with D-glucose-2-phosphate (G2P) | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate binding protein (Agrocinopine) | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C6Y
 
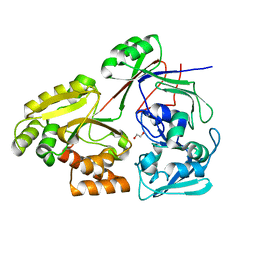 | | PBP AccA from A. tumefaciens Bo542 in apoform 2 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CB9
 
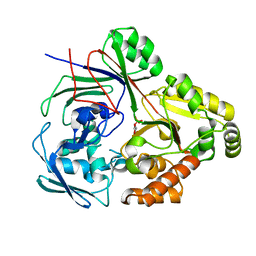 | | PBP AccA from A. tumefaciens Bo542 in complex with D-Glucose-2-phosphate | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-01-25 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
3FKB
 
 | | Structure of NDPK H122G and tenofovir-diphosphate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Morera, S, Chen, Y.X. | | Deposit date: | 2008-12-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleoside diphosphate kinase and the activation of antiviral phosphonate analogs of nucleotides: binding mode and phosphorylation of tenofovir derivatives
Nucleosides Nucleotides Nucleic Acids, 28, 2009
|
|
3IP7
 
 | | Structure of Atu2422-GABA receptor in complex with valine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), CALCIUM ION, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP5
 
 | | Structure of Atu2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP6
 
 | | Structure of Atu2422-GABA receptor in complex with proline | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), PROLINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
