6S5O
 
 | |
3CLP
 
 | | M. loti cyclic-nucleotide binding domain mutant 2 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Mll3241 protein | | Authors: | Clayton, G.M, Altieri, S.L, Thomas, L.R, Morais-Cabral, J.H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Energetic Analysis of Activation by a Cyclic Nucleotide Binding Domain.
J.Mol.Biol., 381, 2008
|
|
3CO2
 
 | |
3CL1
 
 | | M. loti cyclic-nucleotide binding domain, cyclic-GMP bound | | Descriptor: | CHLORIDE ION, CYCLIC GUANOSINE MONOPHOSPHATE, Mll3241 protein, ... | | Authors: | Clayton, G.M, Alteiri, S.L, Thomas, L.R, Morais-Cabral, J.H. | | Deposit date: | 2008-03-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Energetic Analysis of Activation by a Cyclic Nucleotide Binding Domain.
J.Mol.Biol., 381, 2008
|
|
8BZ6
 
 | |
8BZ8
 
 | |
8BZ5
 
 | |
8BZ4
 
 | |
8BZ7
 
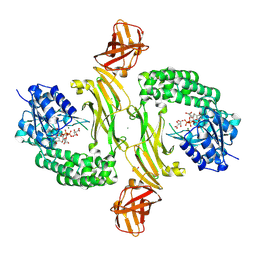 | |
5H9B
 
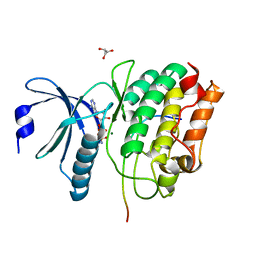 | |
5HIT
 
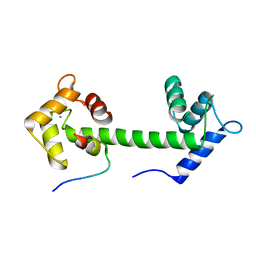 | |
5HU3
 
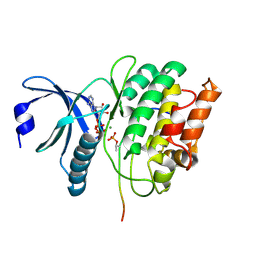 | |
1BEB
 
 | |
1K4C
 
 | | Potassium Channel KcsA-Fab complex in high concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
1K4D
 
 | | Potassium Channel KcsA-Fab complex in low concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
7AGY
 
 | |
7AHT
 
 | |
7AHM
 
 | | Low-resolution structure of the K+/H+ antiporter subunit KhtT in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, K(+)/H(+) antiporter subunit KhtT | | Authors: | Cereija, T.B, Guerra, J.P, Morais-Cabral, J.H. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | c-di-AMP, a likely master regulator of bacterial K + homeostasis machinery, activates a K + exporter.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AGV
 
 | | High-resolution structure of the K+/H+ antiporter subunit KhtT in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETATE ION, CALCIUM ION, ... | | Authors: | Cereija, T.B, Guerra, J.P, Morais-Cabral, J.H. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | c-di-AMP, a likely master regulator of bacterial K + homeostasis machinery, activates a K + exporter.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AGW
 
 | |
6S5D
 
 | |
6S5G
 
 | |
6S5E
 
 | |
6S5B
 
 | |
6S7R
 
 | |
