3HOE
 
 | |
3HOL
 
 | |
1J74
 
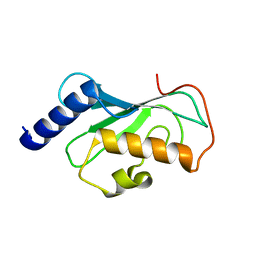 | | Crystal Structure of Mms2 | | Descriptor: | MMS2 | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pastushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
1J7D
 
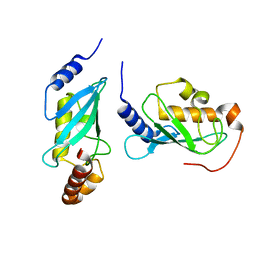 | | Crystal Structure of hMms2-hUbc13 | | Descriptor: | MMS2, UBIQUITIN-CONJUGATING ENZYME E2-17 KDA | | Authors: | Moraes, T.F, Edwards, R.A, McKenna, S, Pashushok, L, Xiao, W, Glover, J.N.M, Ellison, M.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13.
Nat.Struct.Biol., 8, 2001
|
|
3NY7
 
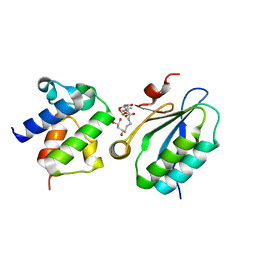 | | STAS domain of YchM bound to ACP | | Descriptor: | 3-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-3-oxopropanoic acid, Acyl carrier protein, GLYCEROL, ... | | Authors: | Moraes, T.F, Reithmeier, R, Strynadka, N.C.S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structure of a SLC26 Anion Transporter STAS Domain in Complex with Acyl Carrier Protein: Implications for E. coli YchM in Fatty Acid Metabolism.
Structure, 18, 2010
|
|
2O4V
 
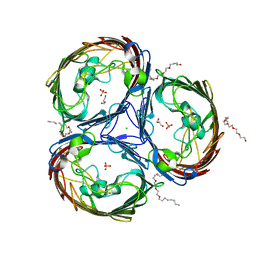 | | An arginine ladder in OprP mediates phosphate specific transfer across the outer membrane | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, T.F, Bains, M, Hancock, R.E, Strynadka, N.C. | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | An arginine ladder in OprP mediates phosphate-specific transfer across the outer membrane.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6N05
 
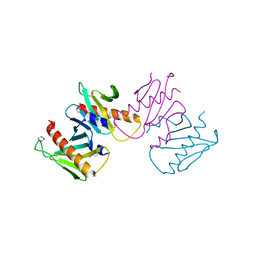 | | Structure of anti-crispr protein, AcrIIC2 | | Descriptor: | AcrIIC2 | | Authors: | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2018-11-06 | | Release date: | 2019-06-05 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
8F3K
 
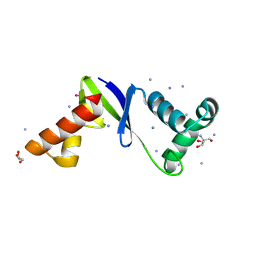 | | Anti-CRISPR protein AcrIIC5 inhibits CRISPR-Cas9 by acting as a DNA mimic | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACRIIC5Nch, AMMONIUM ION, ... | | Authors: | Shah, M, Sungwon, H, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anti-CRISPR Protein AcrIIC5 Inhibits CRISPR-Cas9 by Occupying the Target DNA Binding Pocket.
J.Mol.Biol., 435, 2023
|
|
6WCE
 
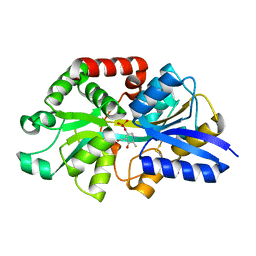 | | Structure of the periplasmic binding protein P5PA | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal-5-phosphate binding protein A (P5PA) | | Authors: | Pan, C, Zimmer, A, Shah, M, Huynh, M, Lai, C.C.L, Sit, B, Hooda, Y, Moraes, T.F. | | Deposit date: | 2020-03-30 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Actinobacillus utilizes a binding protein-dependent ABC transporter to acquire the active form of vitamin B 6 .
J.Biol.Chem., 297, 2021
|
|
4KY9
 
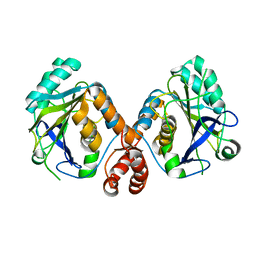 | |
7MSL
 
 | |
8GLO
 
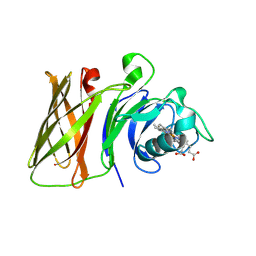 | |
8GM3
 
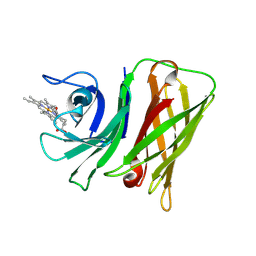 | | Vibrio harveyi Holo HphA | | Descriptor: | HEME B/C, Hemophilin | | Authors: | Pan, C, Shah, M, Moraes, T.F. | | Deposit date: | 2023-03-24 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Heme Acquisition by Slam-dependent Hemophores in Gram-negative Bacteria
To Be Published
|
|
8GMM
 
 | |
6ARZ
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | BROMIDE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Calmettes, C, Shah, M, Pawluk, A, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
4RDR
 
 | |
4R72
 
 | | Structure of the periplasmic binding protein AfuA from Actinobacillus pleuropneumoniae (apo form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC-type Fe3+ transport system, periplasmic component, ... | | Authors: | Sit, B, Calmettes, C, Moraes, T.F. | | Deposit date: | 2014-08-26 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen.
Plos Pathog., 11, 2015
|
|
4R75
 
 | | Structure of the periplasmic binding protein AfuA from Actinobacillus pleuropneumoniae (exogenous sedoheptulose-7-phosphate bound) | | Descriptor: | 1-C-(hydroxymethyl)-6-O-phosphono-beta-D-altrofuranose, ABC-type Fe3+ transport system, periplasmic component, ... | | Authors: | Sit, B, Calmettes, C, Moraes, T.F. | | Deposit date: | 2014-08-26 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen.
Plos Pathog., 11, 2015
|
|
4RDT
 
 | | Structure of the bacterial Zn-transporter ZnuD from Neisseria meningitidis (flexible conformation bound to a zinc ion) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, GLYCEROL, SULFATE ION, ... | | Authors: | Calmettes, C, El Bakkouri, M, Moraes, T.F. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The molecular mechanism of Zinc acquisition by the neisserial outer-membrane transporter ZnuD.
Nat Commun, 6, 2015
|
|
4R74
 
 | | Structure of the periplasmic binding protein AfuA from Actinobacillus pleuropneumoniae (exogenous fructose-6-phosphate bound) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-fructofuranose, ABC-type Fe3+ transport system, ... | | Authors: | Sit, B, Calmettes, C, Moraes, T.F. | | Deposit date: | 2014-08-26 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen.
Plos Pathog., 11, 2015
|
|
4RVW
 
 | |
3VE1
 
 | |
3VE2
 
 | |
6AS4
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 anti-crispr protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
6AS3
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
