2B3T
 
 | | Structure of complex between E. coli translation termination factor RF1 and the PrmC methyltransferase | | Descriptor: | Peptide chain release factor 1, Protein methyltransferase hemK, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Graille, M, Heurgue-Hamard, V, Champ, S, Mora, L, Scrima, N, Ulryck, N, van Tilbeurgh, H, Buckingham, R.H. | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis for bacterial class I release factor methylation by PrmC
Mol.Cell, 20, 2005
|
|
1V74
 
 | | Structure of the E. coli colicin D bound to its immunity protein ImmD | | Descriptor: | Colicin D, Colicin D immunity protein, PENTAETHYLENE GLYCOL | | Authors: | Graille, M, Mora, L, Buckingham, R.H, van Tilbeurgh, H, de Zamaroczy, M. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein
Embo J., 23, 2004
|
|
3Q87
 
 | | Structure of E. cuniculi Mtq2-Trm112 complex responible for the methylation of eRF1 translation termination factor | | Descriptor: | N6 adenine specific DNA methylase, Putative uncharacterized protein ECU08_1170, S-ADENOSYLMETHIONINE, ... | | Authors: | Liger, D, Mora, L, Lazar, N, Figaro, S, Henri, J, Scrima, N, Buckingham, R.H, van Tilbeurgh, H, Heurgue-Hamard, V, Graille, M. | | Deposit date: | 2011-01-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Mechanism of activation of methyltransferases involved in translation by the Trm112 'hub' protein
Nucleic Acids Res., 2011
|
|
7T7W
 
 | |
9BWI
 
 | | Crystal structure of cellulose oxidative enzyme in acidic pH with glycerol | | Descriptor: | COPPER (II) ION, Cellulose oxidative enzyme, GLYCEROL | | Authors: | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
9BWH
 
 | | Crystal structure of cellulose oxidative enzyme with glycerol | | Descriptor: | COPPER (II) ION, Cellulose oxidative enzyme, GLYCEROL | | Authors: | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
9BWF
 
 | | Crystal structure of cellulose oxidative enzyme without ligand | | Descriptor: | COPPER (II) ION, Cellulose oxidative enzyme | | Authors: | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
3HDL
 
 | | Crystal Structure of Highly Glycosylated Peroxidase from Royal Palm Tree | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Watanabe, L, Moura, P.R, Bleicher, L, Nascimento, A.S, Zamorano, L.S, Calvete, J.J, Bursakov, S, Roig, M.G, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2009-05-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and statistical coupling analysis of highly glycosylated peroxidase from royal palm tree (Roystonea regia).
J.Struct.Biol., 169, 2010
|
|
9C1B
 
 | | Crystal structure of GDP-bound human M-RAS protein in crystal form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bester, S.M, Abrahamsen, R, Samora, L.R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the GDP-bound human M-RAS protein in two crystal forms.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6NR6
 
 | | Crystal Structure of Staphylococcus pseudintermedius SbnI in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional transcriptional regulator/O-phospho-L-serine synthase SbnI, MAGNESIUM ION | | Authors: | Verstraete, M.M, Morales, L.D, Kobylarz, M.J, Loutet, S.A, Laakso, H.A, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2019-01-22 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The heme-sensitive regulator SbnI has a bifunctional role in staphyloferrin B production by Staphylococcus aureus .
J.Biol.Chem., 294, 2019
|
|
9C1A
 
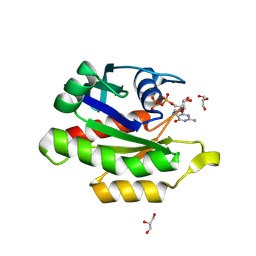 | | Crystal structure of GDP-bound human M-RAS protein in crystal form I | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bester, S.M, Abrahamsen, R, Samora, L.R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the GDP-bound human M-RAS protein in two crystal forms.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
