7WUE
 
 | | Crystal structure of SARS-CoV-2 Receptor Binding Domain in complex with the monoclonal antibody m31A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mohapatra, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models.
Sci Transl Med, 14, 2022
|
|
7YCL
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with IS-9A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, IS-9A Fab light chain, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCN
 
 | |
8HHY
 
 | | SARS-CoV-2 Delta Spike in complex with IS-9A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, ... | | Authors: | Mohapatra, A, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8X0X
 
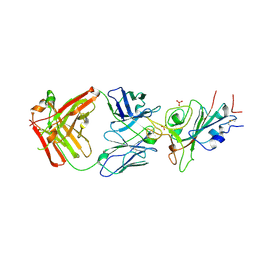 | | Crystal structure of JE-5C in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JE-5C Fab, Light chain of JE-5C Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8X0Y
 
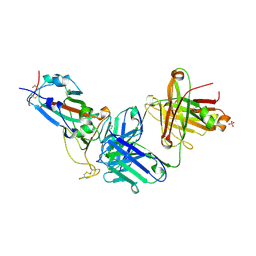 | | Crystal structure of JM-1A in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of JM-1A Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8HHZ
 
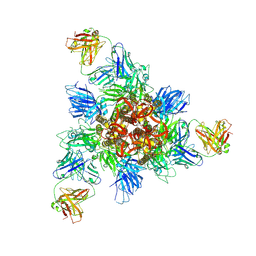 | | SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A | | Descriptor: | IY-2A Fab heavy chain, IY-2A Fab light chain, Spike glycoprotein | | Authors: | Chen, X, Mohapatra, A, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
