5AZD
 
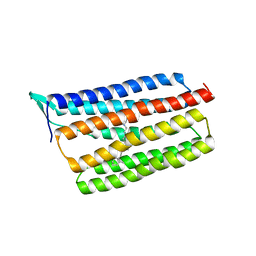 | | Crystal structure of thermophilic rhodopsin. | | Descriptor: | Bacteriorhodopsin | | Authors: | Mizutani, K, Hashimoto, N, Tsukamoto, T, Yamashita, K, Yamamoto, M, Sudo, Y, Murata, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic structure of thermophilic rhodopsin: implications for high thermal stability and optogenetic availability.
To Be Published
|
|
1NFT
 
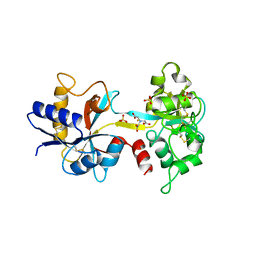 | | OVOTRANSFERRIN, N-TERMINAL LOBE, IRON LOADED OPEN FORM | | Descriptor: | FE (III) ION, NITRILOTRIACETIC ACID, PROTEIN (OVOTRANSFERRIN), ... | | Authors: | Mizutani, K, Yamashita, H, Kurokawa, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1IQ7
 
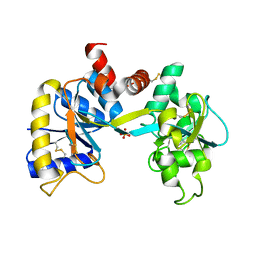 | | Ovotransferrin, C-Terminal Lobe, Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ovotransferrin, SULFATE ION | | Authors: | Mizutani, K, Muralidhara, B.K, Yamashita, H, Tabata, S, Mikami, B, Hirose, M. | | Deposit date: | 2001-07-06 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anion-mediated Fe3+ release mechanism in ovotransferrin C-lobe: a structurally identified SO4(2-) binding site and its implications for the kinetic pathway.
J.Biol.Chem., 276, 2001
|
|
1IEJ
 
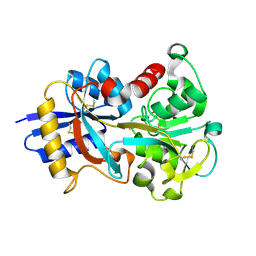 | | OVOTRANSFERRIN, N-TERMINAL LOBE, HOLO FORM, AT 1.65 A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, OVOTRANSFERRIN | | Authors: | Mizutani, K, Mikami, B, Hirose, M. | | Deposit date: | 2001-04-10 | | Release date: | 2001-06-20 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Domain closure mechanism in transferrins: new viewpoints about the hinge structure and motion as deduced from high resolution crystal structures of ovotransferrin N-lobe.
J.Mol.Biol., 309, 2001
|
|
1TFA
 
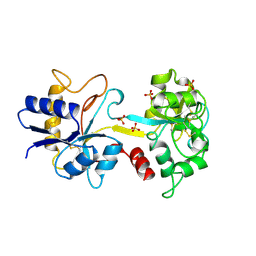 | | OVOTRANSFERRIN, N-TERMINAL LOBE, APO FORM | | Descriptor: | PROTEIN (OVOTRANSFERRIN), SULFATE ION | | Authors: | Mizutani, K, Yamashita, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1V9F
 
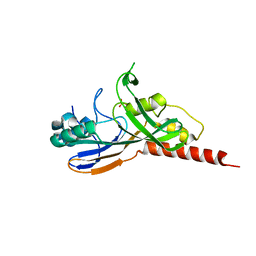 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
3AOU
 
 | | Structure of the Na+ unbound rotor ring modified with N,N f-Dicyclohexylcarbodiimide of the Na+-transporting V-ATPase | | Descriptor: | DICYCLOHEXYLUREA, UNDECYL-MALTOSIDE, V-type sodium ATPase subunit K | | Authors: | Mizutani, K, Yamamoto, M, Yamato, I, Kakinuma, Y, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ABG
 
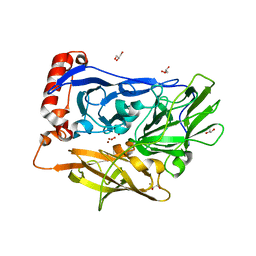 | | X-ray Crystal Analysis of Bilirubin Oxidase from Myrothecium verrucaria at 2.3 angstrom Resolution using a Twin Crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Mizutani, K, Toyoda, M, Sagara, K, Takahashi, N, Sato, A, Kamitaka, Y, Tsujimura, S, Nakanishi, Y, Sugiura, T, Yamaguchi, S, Kano, K, Mikami, B. | | Deposit date: | 2009-12-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 A resolution using a twinned crystal
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3VUP
 
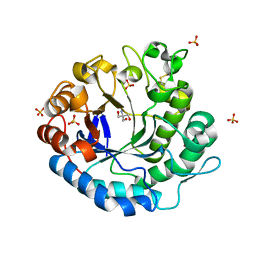 | | Beta-1,4-mannanase from the common sea hare Aplysia kurodai | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase, SULFATE ION | | Authors: | Mizutani, K, Tsuchiya, S, Toyoda, M, Nanbu, Y, Tominaga, K, Yuasa, K, Takahashi, N, Tsuji, A, Mikami, B. | | Deposit date: | 2012-07-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of beta-1,4-mannanase from the common sea hare Aplysia kurodai at 1.05 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3VM5
 
 | |
2D3I
 
 | | Crystal Structure of Aluminum-Bound Ovotransferrin at 2.15 Angstrom Resolution | | Descriptor: | ALUMINUM ION, BICARBONATE ION, Ovotransferrin | | Authors: | Mizutani, K, Mikami, B, Aibara, S, Hirose, M. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of aluminium-bound ovotransferrin at 2.15 Angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8X3H
 
 | |
8Y1M
 
 | | Xylanase R from Bacillus sp. TAR-1 complexed with xylobiose. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Nakamura, T, Kuwata, K, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-25 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8XY0
 
 | | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase A | | Authors: | Nakamura, T, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8YWO
 
 | | Crystal structure of L-azetidine-2-carboxylate hydrolase soaked in (S)-azetidine-2-carboxylic acid | | Descriptor: | (2S)-azetidine-2-carboxylic acid, (S)-2-haloacid dehalogenase | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-31 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
8YVW
 
 | | Crystal structure of D12N mutant of L-azetidine-2-carboxylate hydrolase | | Descriptor: | (S)-2-haloacid dehalogenase, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
9JF5
 
 | | Arginine decarboxylase in Aspergillus oryzae complexed with arginine | | Descriptor: | 1,2-ETHANEDIOL, AGMATINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9JFN
 
 | | Arginine decarboxylase in Aspergillus oryzae complexed with agmatine | | Descriptor: | 1,2-ETHANEDIOL, AGMATINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-05 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9JER
 
 | | Arginine decarboxylase in Aspergillus oryzae, ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-tryptophan decarboxylase PsiD-like domain-containing protein | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
5CL2
 
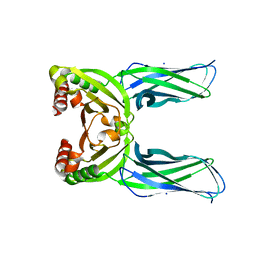 | | Crystal structure of Spo0M, sporulation control protein, from Bacillus subtilis. | | Descriptor: | SODIUM ION, Sporulation-control protein spo0M | | Authors: | Sonoda, Y, Mizutani, K, Mikami, B. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Spo0M, a sporulation-control protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2OHE
 
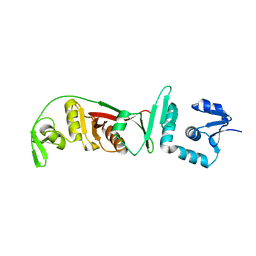 | | Structural and mutational analysis of tRNA-Intron splicing endonuclease from Thermoplasma acidophilum DSM 1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
2OHC
 
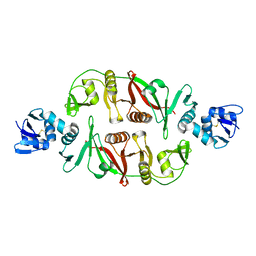 | | structural and mutational analysis of tRNA-intron splicing endonuclease from Thermoplasma acidophilum DSM1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
5KNC
 
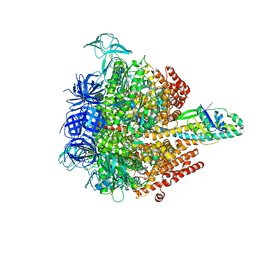 | | Crystal structure of the 3 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KNB
 
 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KND
 
 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
