8JPW
 
 | | Crystal Structure of Single-chain L-Glutamate Oxidase Mutant from Streptomyces sp. X-119-6 | | Descriptor: | 2-OXOGLUTARIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase, ... | | Authors: | Yamaguchi, H, Takahashi, K, Tatsumi, M, Tagami, U, Mizukoshi, T, Miyano, H, Sugiki, M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Development of a novel single-chain l-glutamate oxidase from Streptomyces sp. X-119-6 by inserting flexible linkers.
Enzyme.Microb.Technol., 170, 2023
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | Descriptor: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | Authors: | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
2KSV
 
 | | The NMR structure of protein-glutaminase from Chryseobacterium proteolyticum | | Descriptor: | Protein-glutaminase | | Authors: | Kumeta, H, Miwa, N, Ogura, K, Kai, Y, Mizukoshi, T, Shimba, N, Suzuki, E, Inagaki, F. | | Deposit date: | 2010-01-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of protein-glutaminase from Chryseobacterium proteolyticum.
J.Biomol.Nmr, 46, 2010
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | Descriptor: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D48
 
 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | Descriptor: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
7C8H
 
 | | Ambient temperature structure of Bifidobacterium longum phosphoketolase with thiamine diphosphate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, MALONIC ACID, ... | | Authors: | Nakata, K, Kashiwagi, T, Nango, E, Miyano, H, Mizukoshi, T, Iwata, S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7C8I
 
 | | Ambient temperature structure of Bifidobacgterium longum phosphoketolase with thiamine diphosphate and phosphoenol pyuruvate | | Descriptor: | CALCIUM ION, PHOSPHOENOLPYRUVATE, THIAMINE DIPHOSPHATE, ... | | Authors: | Nakata, K, Kashiwagi, T, Nango, E, Miyano, H, Mizukoshi, T, Iwata, S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5ZBC
 
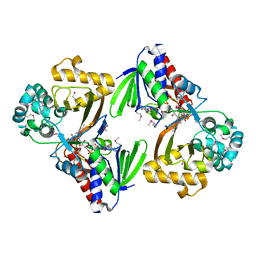 | | Crystal structure of Se-Met tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | Authors: | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | Deposit date: | 2018-02-11 | | Release date: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
5ZBD
 
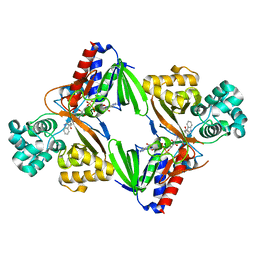 | | Crystal structure of tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, TRYPTOPHAN | | Authors: | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | Deposit date: | 2018-02-11 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
6ACF
 
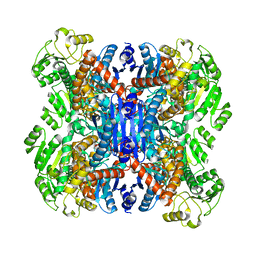 | | structure of leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
6ACH
 
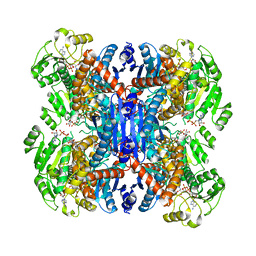 | | Structure of NAD+-bound leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
7ERV
 
 | | Crystal structure of L-histidine decarboxylase (C57S/C101V/C282V mutant) from Photobacterium phosphoreum | | Descriptor: | Histidine decarboxylase, IMIDAZOLE | | Authors: | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
7ERU
 
 | | Crystal structure of L-histidine decarboxylase (C57S mutant) from Photobacterium phosphoreum | | Descriptor: | Histidine decarboxylase | | Authors: | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
