3LZ8
 
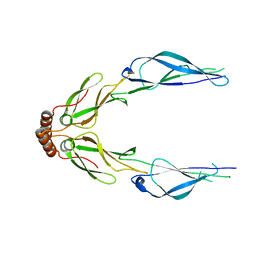 | | Structure of a putative chaperone dnaj from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 2.9 a resolution. | | Descriptor: | Putative chaperone DnaJ | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Bearden, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Putative Chaperone Dnaj from Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578 at 2.9 A Resolution.
To be Published
|
|
3MAH
 
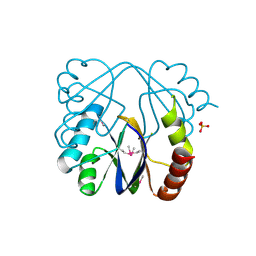 | | A putative c-terminal regulatory domain of aspartate kinase from porphyromonas gingivalis w83. | | Descriptor: | Aspartokinase, SULFATE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Moy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Putative C-Terminal Regulatory Domain of Aspartate Kinase from Porphyromonas Gingivalis W83.
To be Published
|
|
3NVT
 
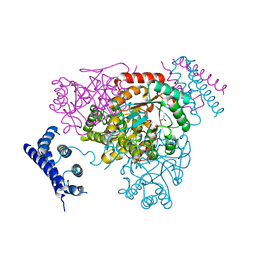 | | 1.95 Angstrom crystal structure of a bifunctional 3-deoxy-7-phosphoheptulonate synthase/chorismate mutase (aroA) from Listeria monocytogenes EGD-e | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, ACETATE ION, MANGANESE (II) ION | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain.
Protein Sci., 21, 2012
|
|
3V4G
 
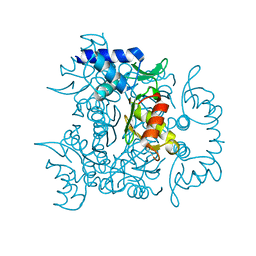 | | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6 | | Descriptor: | Arginine repressor | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
To be Published
|
|
4ISC
 
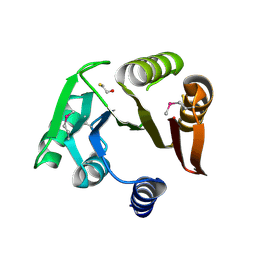 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
5UG4
 
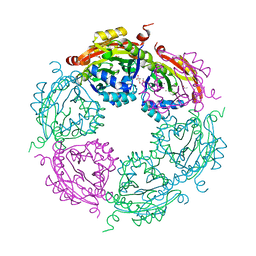 | | Structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-06 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae
To Be Published
|
|
5UPR
 
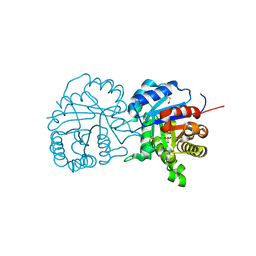 | | X-ray structure of a putative triosephosphate isomerase from Toxoplasma gondii ME49 | | Descriptor: | CHLORIDE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Cardona-Correa, A, Bishop, B, Anderson, W.F, Ngo, H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a putative triosephosphate isomerase from Toxoplasma gondii ME49
To Be Published
|
|
3QFG
 
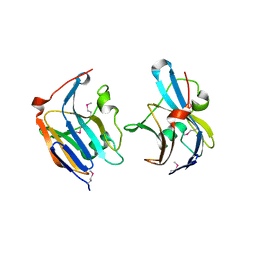 | | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Halavaty, A, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
4JM7
 
 | | 1.82 Angstrom resolution crystal structure of holo-(acyl-carrier-protein) synthase (acpS) from Staphylococcus aureus | | Descriptor: | Holo-[acyl-carrier-protein] synthase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4JNN
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine | | Descriptor: | BENZAMIDINE, BETA-MERCAPTOETHANOL, Transcriptional regulator | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-15 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine
To be Published
|
|
3R2I
 
 | | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Exotoxin 5 | | Authors: | Filippova, E.V, Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3TK7
 
 | | 2.0 Angstrom Resolution Crystal Structure of Transaldolase B (TalA) from Francisella tularensis in Covalent Complex with Fructose 6-Phosphate | | Descriptor: | FRUCTOSE -6-PHOSPHATE, Transaldolase | | Authors: | Light, S.H, Minasov, G, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-25 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adherence to Burgi-Dunitz stereochemical principles requires significant structural rearrangements in Schiff-base formation: insights from transaldolase complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3RU6
 
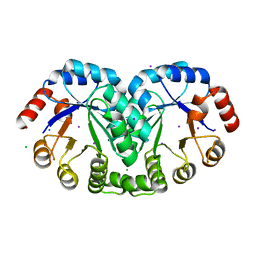 | | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, IODIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4M8I
 
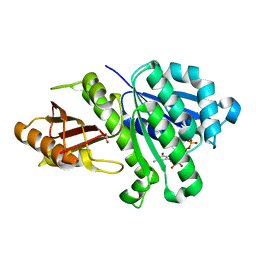 | | 1.43 Angstrom resolution crystal structure of cell division protein FtsZ (ftsZ) from Staphylococcus epidermidis RP62A in complex with GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Filippova, E.V, Olsen, D.B, Therien, A, Shuvalova, L, Young, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-13 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | 1.43 Angstrom resolution crystal structure of cell division protein FtsZ (ftsZ) from Staphylococcus epidermidis RP62A in complex with GDP
To be Published
|
|
3S42
 
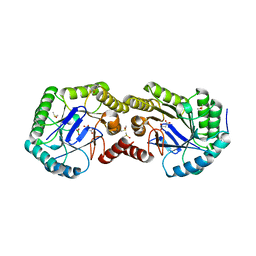 | | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site | | Descriptor: | 3-dehydroquinate dehydratase, BORIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Halavaty, A.S, Krishna, S.N, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site
To be Published
|
|
3NQR
 
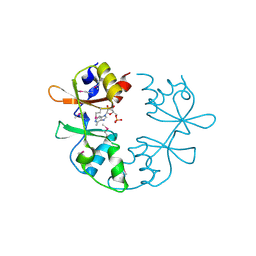 | | A putative CBS domain-containing protein from Salmonella typhimurium LT2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein corC | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Cui, H, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A putative CBS domain-containing protein from Salmonella typhimurium LT2.
To be Published
|
|
5TR7
 
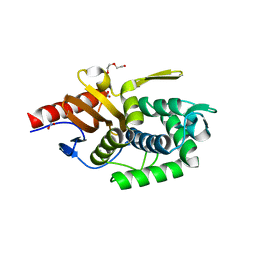 | | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961
To Be Published
|
|
5TXU
 
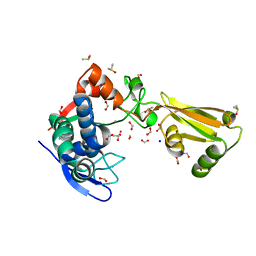 | | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Nocadello, S, Minasov, G, Kiryukhina, O, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation
To Be Published
|
|
5UE1
 
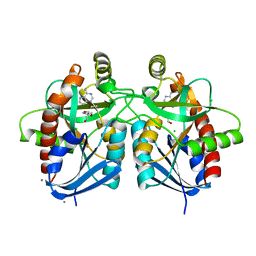 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with adenine from Vibrio fischeri ES114 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Vibrio fischeri ES114
To Be Published
|
|
3OGA
 
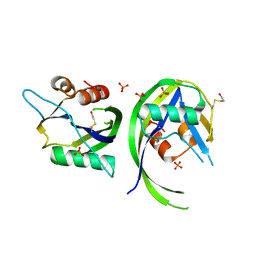 | | 1.75 Angstrom resolution crystal structure of a putative NTP pyrophosphohydrolase (yfaO) from Salmonella typhimurium LT2 | | Descriptor: | BETA-MERCAPTOETHANOL, Nucleoside triphosphatase nudI, PHOSPHATE ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of a putative NTP pyrophosphohydrolase (yfaO) from Salmonella typhimurium LT2
To be Published
|
|
3R38
 
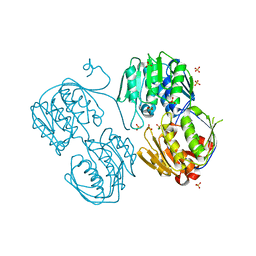 | | 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, SULFATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-15 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e
To be Published
|
|
3ROI
 
 | | 2.20 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, CHLORIDE ION, SULFATE ION | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.20 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii
To be Published
|
|
3OO2
 
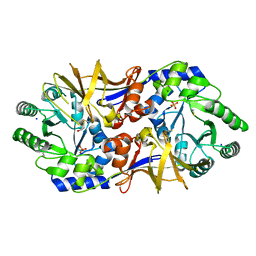 | | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Alanine racemase 1, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3SC6
 
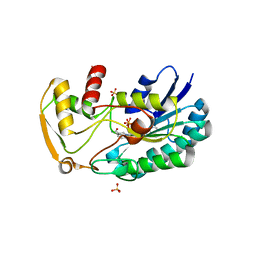 | | 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, dTDP-4-dehydrorhamnose reductase | | Authors: | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-07 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose reductase (RfbD).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3S83
 
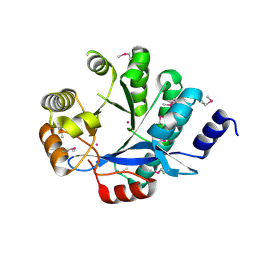 | | EAL domain of phosphodiesterase PdeA | | Descriptor: | GGDEF family protein, POTASSIUM ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of EAL domain from Caulobacter crescentus CB15
To be Published
|
|
