4JRH
 
 | | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio Cholerae (space group P43) at 2.2 Angstrom | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, ACETATE ION | | Authors: | Hou, J, Chruszcz, M, Shabalin, I.G, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: |
|
|
4QGM
 
 | | Acireductone dioxygenase from Bacillus anthracis with cadmium ion in active center | | Descriptor: | Acireductone dioxygenase, CADMIUM ION | | Authors: | Milaczewska, A.M, Chruszcz, M, Shabalin, I.G, Cooper, D.R, Borowski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Acireductone dioxygenase from Bacillus anthracis with cadmium ion in active center
To be Published
|
|
3TNJ
 
 | | Crystal structure of universal stress protein from Nitrosomonas europaea with AMP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, Universal stress protein (Usp) | | Authors: | Tkaczuk, K.L, Chruszcz, M, Shumilin, I.A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
2FEX
 
 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | Authors: | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
2FI0
 
 | | The crystal structure of the conserved domain protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | conserved domain protein | | Authors: | Zhang, R, Li, H, Abdullah, J, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-27 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the conserved domain protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
2ARK
 
 | | Structure of a flavodoxin from Aquifex aeolicus | | Descriptor: | Flavodoxin, GLYCEROL, PHOSPHATE ION | | Authors: | Cuff, M.E, Quartey, P, Zhou, M, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a flavodoxin from Aquifex aeolicus
To be Published
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
2FI9
 
 | | The crystal structure of an outer membrane protein from the Bartonella henselae | | Descriptor: | Outer membrane protein | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-28 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8A crystal structure of an outer membrane protein from the Bartonella henselae
To be Published
|
|
1IW2
 
 | | X-ray structure of Human Complement Protein C8gamma at pH=7.O | | Descriptor: | Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
2QMO
 
 | | Crystal structure of dethiobiotin synthetase (bioD) from Helicobacter pylori | | Descriptor: | CHLORIDE ION, Dethiobiotin synthetase | | Authors: | Chruszcz, M, Xu, X, Cuff, M, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-16 | | Release date: | 2007-07-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
2FHX
 
 | | Pseudomonas aeruginosa SPM-1 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Murphy, T.A, Catto, L.E, Halford, S.E, Hadfield, A.T, Minor, W, Walsh, T.R, Spencer, J. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pseudomonas aeruginosa SPM-1 Provides Insights into Variable Zinc Affinity of Metallo-beta-lactamases.
J.Mol.Biol., 357, 2006
|
|
4OMR
 
 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, BENZAMIDINE, Enoyl-CoA hydratase | | Authors: | Langner, K.M, Cooper, D.R, Chapman, H.C, Handing, K.B, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-27 | | Release date: | 2014-04-02 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA
To be Published
|
|
4O9K
 
 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
2QNU
 
 | | Crystal structure of PA0076 from Pseudomonas aeruginosa PAO1 at 2.05 A resolution | | Descriptor: | ACETATE ION, TRIETHYLENE GLYCOL, Uncharacterized protein PA0076 | | Authors: | Filippova, E.V, Chruszcz, M, Skarina, T, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Pa0076 from Pseudomonas aeruginosa PAO1 at 2.05 A resolution.
To be Published
|
|
2G3A
 
 | | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | acetyltransferase | | Authors: | Cymborowski, M, Xu, X, Chruszcz, M, Zheng, H, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | Descriptor: | GLYCEROL, putative isochorismatase hydrolase | | Authors: | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
7KYJ
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Shabalin, I.G, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with zinc
To Be Published
|
|
1LF7
 
 | | Crystal Structure of Human Complement Protein C8gamma at 1.2 A Resolution | | Descriptor: | CITRIC ACID, Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
6BII
 
 | | Crystal Structure of Pyrococcus yayanosii Glyoxylate Hydroxypyruvate Reductase in complex with NADP and malonate (re-refinement of 5AOW) | | Descriptor: | GLYCEROL, Glyoxylate reductase, MALONATE ION, ... | | Authors: | Lassalle, L, Shabalin, I.G, Girard, E, Minor, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases.
Sci Rep, 6, 2016
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3M3P
 
 | | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus | | Descriptor: | Glutamine amido transferase | | Authors: | Fedorov, A.A, Domagalski, M, Fedorov, E.V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus
To be Published
|
|
4OAE
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol
To be Published
|
|
4OG1
 
 | | Crystal Structure of a Putative Enoyl-CoA Hydratase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Enoyl-CoA hydratase/isomerase | | Authors: | Chapman, H.C, Cooper, D.R, Tkaczuk, K.L, Cymborowski, M.T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Alkire, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of a Putative Enoyl-CoA Hydratase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4POZ
 
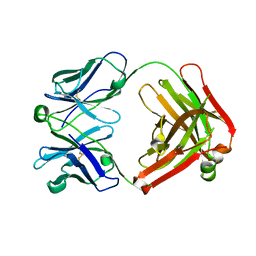 | | Fab fragment of Der p 1 specific antibody 10B9 | | Descriptor: | heavy chain of Fab fragment of 10B9 antibody, light chain of Fab fragment of 10B9 antibody | | Authors: | Osinski, T, Majorek, K.A, Pomes, A, Offermann, L.R, Osinski, S, Glesner, J, Vailes, L.D, Chapman, M.D, Minor, W, Chruszcz, M. | | Deposit date: | 2014-02-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Der p 1-Antibody Complexes and Comparison with Complexes of Proteins or Peptides with Monoclonal Antibodies.
J. Immunol., 195, 2015
|
|
