8OYD
 
 | |
3BC9
 
 | | Alpha-amylase B in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3BCF
 
 | | Alpha-amylase B from Halothermothrix orenii | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
7FGL
 
 | | The complex structure of PHF core domain peptide of tau, VQIVYK, and antibody's Fab domain. | | Descriptor: | AMMONIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cross-reaction complex structure with VQIVYK of tau and the antibody's Fab domain.
To Be Published
|
|
6HJ7
 
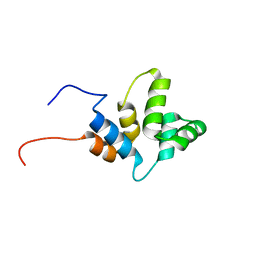 | |
4EXO
 
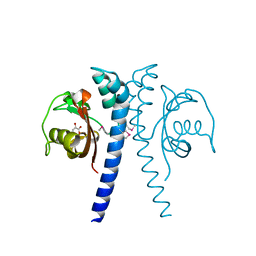 | | Revised, rerefined crystal structure of PDB entry 2QHK, methyl accepting chemotaxis protein | | Descriptor: | Methyl-accepting chemotaxis protein, PYRUVIC ACID | | Authors: | Sweeney, E.G, Henderson, J.N, Goers, J, Wreden, C, Hicks, K.G, Foster, J.K, Parthasarathy, R, Remington, S.J, Guillemin, K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Proposed Mechanism for the pH-Sensing Helicobacter pylori Chemoreceptor TlpB.
Structure, 20, 2012
|
|
8CM3
 
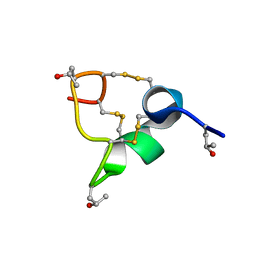 | |
6DTM
 
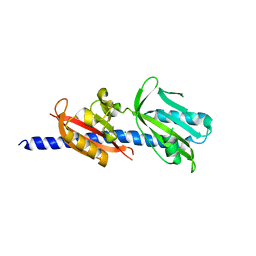 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein TlpA | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-06-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
6E0A
 
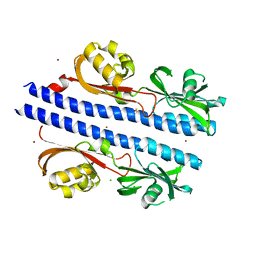 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-07-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
6E09
 
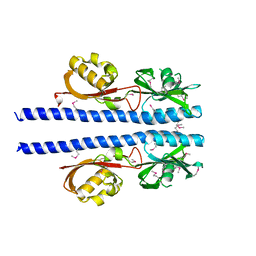 | |
3B23
 
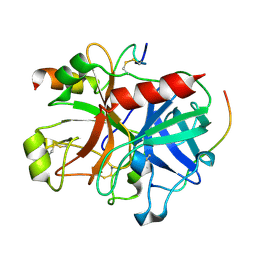 | | Crystal structure of thrombin-variegin complex: Insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors | | Descriptor: | Thrombin heavy chain, Thrombin light chain, Variegin | | Authors: | Koh, C.Y, Kumar, S, Swaminathan, K, Kini, R.M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with s-variegin: insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors
Plos One, 6, 2011
|
|
5DTO
 
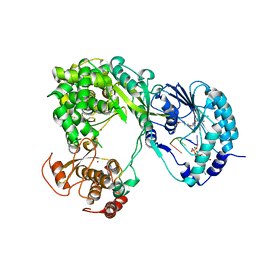 | | Dengue virus full length NS5 complexed with viral Cap 0-RNA and SAH | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Soh, T.S, Lim, S.P, Chung, K.Y, Swaminathan, K, Vasudevan, S.G, Shi, P.-Y, Lescar, J, Luo, D. | | Deposit date: | 2015-09-18 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5)
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7QZV
 
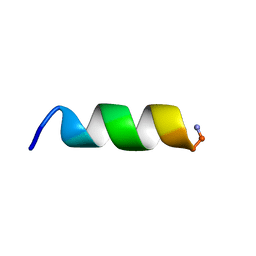 | | Hm-AMP2 | | Descriptor: | Hm-AMP2 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Non-toxic antimicrobial peptide Hm-AMP2 from leech metagenome proteins identified by the gradient-boosting approach
Materials, 224, 2022
|
|
7QZW
 
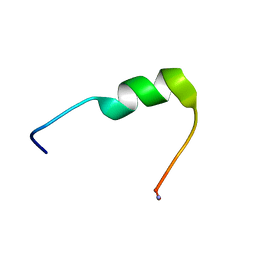 | | Hm-AMP8 | | Descriptor: | Hm-AMP8 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | The study of the interaction of cationic peptides from a medical leech with blood components as a first step to the systemic use of antimicrobial peptides for therapy of infectious diseases
To Be Published
|
|
5ZTZ
 
 | |
8UZW
 
 | |
3UB9
 
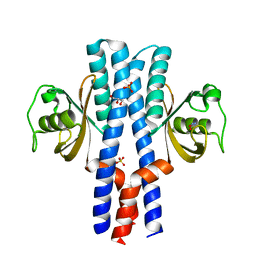 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with hydroxyurea bound | | Descriptor: | GLYCEROL, N-HYDROXYUREA, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
7W7R
 
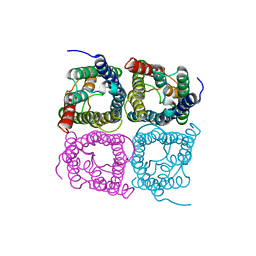 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7W7S
 
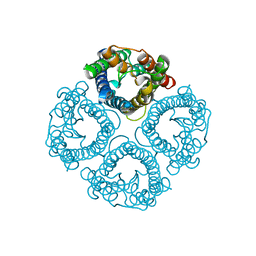 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
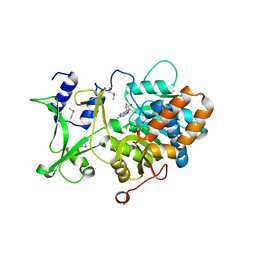
5ZU0
 
 | |
5ZZW
 
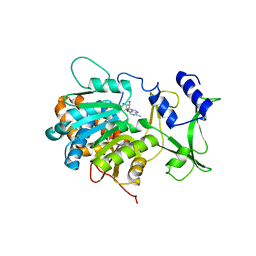 | |
7VTJ
 
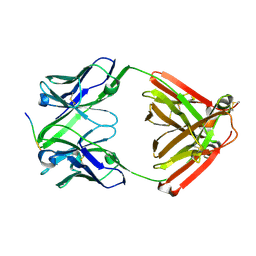 | | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain. | | Descriptor: | Heavy chain of Fab, Light chain of Fab, VQIIYK peptide | | Authors: | Tsuchida, T, Fukuhara, N, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, Y, Ishida, T, Tomoo, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain.
To Be Published
|
|
7VNX
 
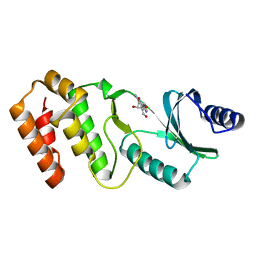 | | Crystal structure of TkArkI | | Descriptor: | GUANOSINE, TkArkI | | Authors: | Yamashita, S, Minowa, K, Ohira, T, Suzuki, T, Tomita, K. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance.
Nature, 605, 2022
|
|
3K1D
 
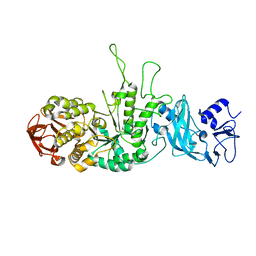 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
3UB7
 
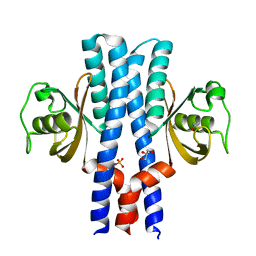 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with acetamide bound | | Descriptor: | ACETAMIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
