5B3P
 
 | |
5B3Q
 
 | |
5B3Y
 
 | |
7E4L
 
 | |
5B3W
 
 | | Crystal structure of hPin1 WW domain (5-15) fused with maltose-binding protein in C2221 form | | Descriptor: | CITRIC ACID, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2016-03-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of the N-terminal fragments of the WW domain: Insights into co-translational folding of a beta-sheet protein
Sci Rep, 6, 2016
|
|
5B3X
 
 | |
5BMY
 
 | |
5AUQ
 
 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUO
 
 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUP
 
 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUN
 
 | | Crystal structure of the HypAB-Ni complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7VLE
 
 | | Oxy-deoxy intermediate of V2 hemoglobin at 55% oxygen saturation | | Descriptor: | Extracellular A1 globin, Extracellular A2 globin, Extracellular B1 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLD
 
 | | Oxy-deoxy intermediate of V2 hemoglobin at 69% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLF
 
 | | Oxy-deoxy intermediate of V2 hemoglobin at 26% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLC
 
 | | Oxy-deoxy intermediate of V2 hemoglobin at 78% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
6JBD
 
 | | Phosphotransferase-ATP complex related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
6JBC
 
 | | Phosphotransferase related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
2D05
 
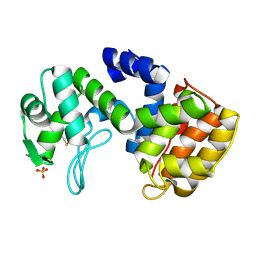 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
1WNR
 
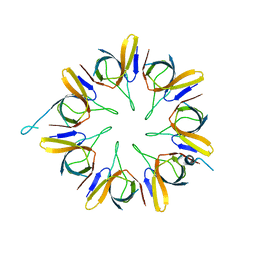 | |
1Z1E
 
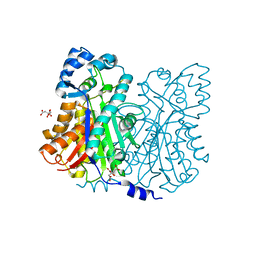 | | Crystal structure of stilbene synthase from Arachis hypogaea | | Descriptor: | CITRIC ACID, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
1Z1F
 
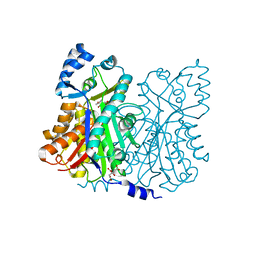 | | Crystal structure of stilbene synthase from Arachis hypogaea (resveratrol-bound form) | | Descriptor: | CITRIC ACID, RESVERATROL, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
1IUC
 
 | | Fucose-specific lectin from Aleuria aurantia with three ligands | | Descriptor: | Fucose-specific lectin, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Fujihashi, M, Peapus, D.H, Kamiya, N, Nagata, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-01 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Fucose-Specific Lectin from Aleuria aurantia Binding Ligands at Three of Its Five Sugar Recognition Sites
Biochemistry, 42, 2003
|
|
1IWM
 
 | | Crystal Structure of the Outer Membrane Lipoprotein Receptor, LolB | | Descriptor: | Outer Membrane Lipoprotein LolB, SULFATE ION | | Authors: | Takeda, K, Miyatake, H, Yokota, N, Matsuyama, S, Tokuda, H, Miki, K. | | Deposit date: | 2002-05-17 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of bacterial lipoprotein localization factors, LolA and LolB.
Embo J., 22, 2003
|
|
1JFL
 
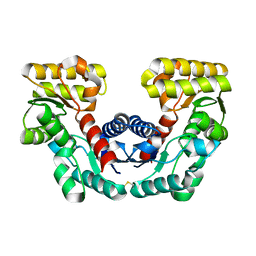 | | CRYSTAL STRUCTURE DETERMINATION OF ASPARTATE RACEMASE FROM AN ARCHAEA | | Descriptor: | ASPARTATE RACEMASE | | Authors: | Liu, L.J, Iwata, K, Kita, A, Kawarabayasi, Y, Yohda, M, Miki, K. | | Deposit date: | 2001-06-21 | | Release date: | 2002-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartate racemase from Pyrococcus horikoshii OT3 and its implications for molecular mechanism of PLP-independent racemization.
J.Mol.Biol., 319, 2002
|
|
1IOK
 
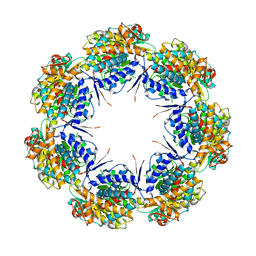 | | CRYSTAL STRUCTURE OF CHAPERONIN-60 FROM PARACOCCUS DENITRIFICANS | | Descriptor: | CHAPERONIN 60 | | Authors: | Fukami, T.A, Yohda, M, Taguchi, H, Yoshida, M, Miki, K. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of chaperonin-60 from Paracoccus denitrificans.
J.Mol.Biol., 312, 2001
|
|
