5YO8
 
 | |
1UD3
 
 | | Crystal structure of AmyK38 N289H mutant | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD8
 
 | | Crystal structure of AmyK38 with lithium ion | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD5
 
 | | Crystal structure of AmyK38 with rubidium ion | | Descriptor: | RUBIDIUM ION, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UA8
 
 | | Crystal structure of the lipoprotein localization factor, LolA | | Descriptor: | Outer-membrane lipoproteins carrier protein | | Authors: | Takeda, K, Miyatake, H, Yokota, N, Matsuyama, S, Tokuda, H, Miki, K. | | Deposit date: | 2003-03-04 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of bacterial lipoprotein localization factors, LolA and LolB.
Embo J., 22, 2003
|
|
1UD4
 
 | | Crystal structure of calcium free alpha amylase from Bacillus sp. strain KSM-K38 (AmyK38, in calcium containing solution) | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1VAT
 
 | | Iodine derivative of hen egg-white lysozyme | | Descriptor: | IODIDE ION, Lysozyme C | | Authors: | Takeda, K, Miyatake, H, Park, S.Y, Kawamoto, M, Kamiya, N, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-19 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Multi-wavelength anomalous diffraction method for I and Xe atoms using ultra-high-energy X-rays from SPring-8
J.Appl.Crystallogr., 37, 2004
|
|
1VAU
 
 | | Xenon derivative of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Takeda, K, Miyatake, H, Park, S.Y, Kawamoto, M, Kamiya, N, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-19 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multi-wavelength anomalous diffraction method for I and Xe atoms using ultra-high-energy X-rays from SPring-8
J.Appl.Crystallogr., 37, 2004
|
|
1UD2
 
 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | Descriptor: | GLYCEROL, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
2D2M
 
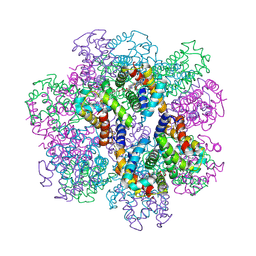 | | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi | | Descriptor: | Giant hemoglobin, A1(b) globin chain, A2(a5) globin chain, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2D2N
 
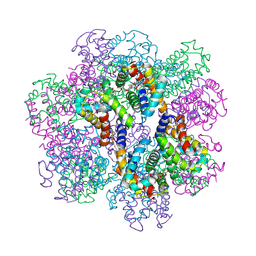 | | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi | | Descriptor: | Giant hemoglobin, A1(b) globin chain, A2(a5) globin chain, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2ECR
 
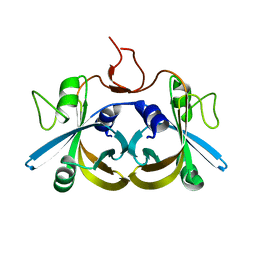 | | Crystal structure of the ligand-free form of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2D81
 
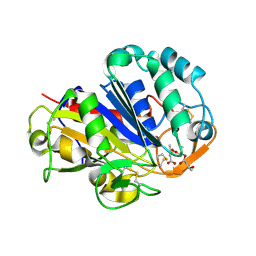 | | PHB depolymerase (S39A) complexed with R3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHB depolymerase | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
2D80
 
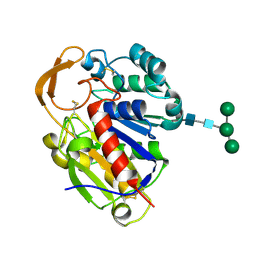 | | Crystal structure of PHB depolymerase from Penicillium funiculosum | | Descriptor: | PHB depolymerase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
1IQR
 
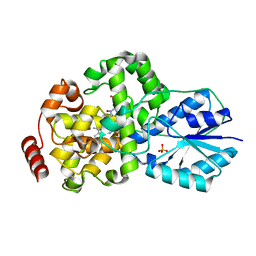 | | Crystal structure of DNA photolyase from Thermus thermophilus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, photolyase | | Authors: | Komori, H, Masui, R, Kuramitsu, S, Yokoyama, S, Shibata, T, Inoue, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-07-27 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IT6
 
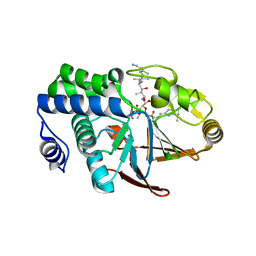 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CALYCULIN A AND THE CATALYTIC SUBUNIT OF PROTEIN PHOSPHATASE 1 | | Descriptor: | CALYCULIN A, MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 1 GAMMA (PP1-GAMMA) CATALYTIC SUBUNIT | | Authors: | Kita, A, Matsunaga, S, Takai, A, Kataiwa, H, Wakimoto, T, Fusetani, N, Isobe, M, Miki, K. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1.
Structure, 10, 2002
|
|
1IQU
 
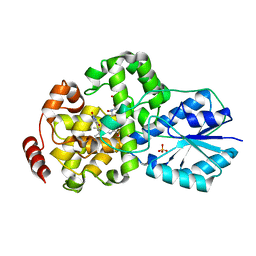 | | Crystal structure of photolyase-thymine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, THYMINE, ... | | Authors: | Komori, H, Masui, R, Kuramitsu, S, Yokoyama, S, Shibata, T, Inoue, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-08-03 | | Release date: | 2002-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5XSW
 
 | |
5YSQ
 
 | | Sulfate-complex structure of a pyrophosphate-dependent kinase in the ribokinase family provides insight into the donor-binding mode | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, SULFATE ION, TM0415 | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
1UBH
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UBU
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
5YSP
 
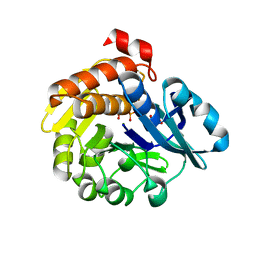 | | Pyrophosphate-dependent kinase in the ribokinase family complexed with a pyrophosphate analog and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, MAGNESIUM ION, METHYLENEDIPHOSPHONIC ACID, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
5ZBY
 
 | |
5YY0
 
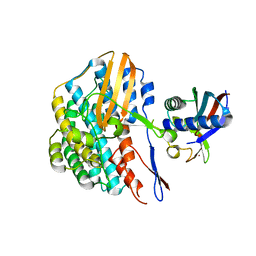 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YXY
 
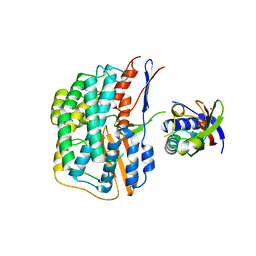 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
