8ZRZ
 
 | |
2E8Z
 
 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with alpha-cyclodextrin | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E9B
 
 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with maltose | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E8Y
 
 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168
in complex with maltose and alpha-cyclodextrin
To be Published
|
|
5Y6Q
 
 | |
4F13
 
 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | Descriptor: | Alginate lyase | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1BTC
 
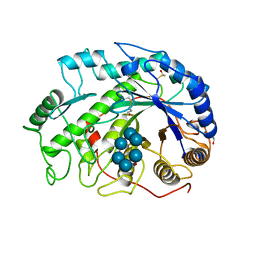 | | THREE-DIMENSIONAL STRUCTURE OF SOYBEAN BETA-AMYLASE DETERMINED AT 3.0 ANGSTROMS RESOLUTION: PRELIMINARY CHAIN TRACING OF THE COMPLEX WITH ALPHA-CYCLODEXTRIN | | Descriptor: | BETA-AMYLASE, BETA-MERCAPTOETHANOL, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Mikami, B, Hehre, E.J, Sato, M, Katsube, Y, Hirose, M, Morita, Y, Sacchettini, J.C. | | Deposit date: | 1993-02-18 | | Release date: | 1993-10-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0-A resolution structure of soybean beta-amylase complexed with alpha-cyclodextrin.
Biochemistry, 32, 1993
|
|
9JI6
 
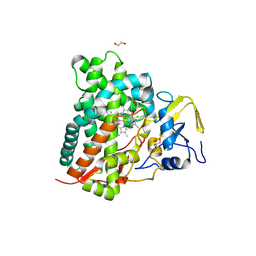 | | CYP105A1 R84A complexed with diclofenac (DIF) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-11 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JIP
 
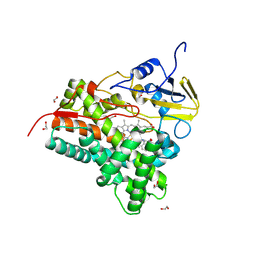 | | CYP105A1 R84A complexed with flufenamic acid (FLF) | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-12 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JHW
 
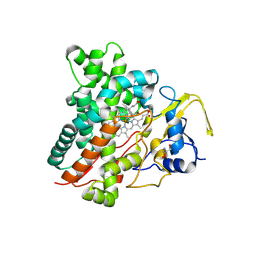 | | CYP105A1 complexed with diclofenac (DIF) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JI1
 
 | | CYP105A1 complexed with flufenamic acid (FLF) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-11 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
5ZU2
 
 | | Effect of mutation (R554A) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
5ZU3
 
 | | Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Formate oxidase, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
9JL5
 
 | | CYP105A1 R84A complexed with mevastatin (cryogenic data collection) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Yasukawa, K, Yoneda, S, Yamagata, M. | | Deposit date: | 2024-09-18 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Room-temperature X-ray data collection enabled the structural determination of statin-bound CYP105A1.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9JKW
 
 | | CYP105A1 R84A complexed with mevastatin | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,8S,8aR)-2-methyl-8-{[(2S)-2-methylbutanoyl]oxy}-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]h eptanoic acid, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D3 dihydroxylase | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Yasukawa, K, Yoneda, S, Yamagata, M. | | Deposit date: | 2024-09-17 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Room-temperature X-ray data collection enabled the structural determination of statin-bound CYP105A1.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9JKZ
 
 | | CYP105A1 R84A complexed with simvastatin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Simvastatin acid, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Yasukawa, K, Yoneda, S, Yamagata, M. | | Deposit date: | 2024-09-17 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room-temperature X-ray data collection enabled the structural determination of statin-bound CYP105A1.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5EEO
 
 | | soybean lipoxygenase(L1)-T756R | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Mikami, B, Nagaya, T, Hioki, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for specificity in lipoxygenase catalysis
To Be Published
|
|
1BYD
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
9JF5
 
 | | Arginine decarboxylase in Aspergillus oryzae complexed with arginine | | Descriptor: | 1,2-ETHANEDIOL, AGMATINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9JER
 
 | | Arginine decarboxylase in Aspergillus oryzae, ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-tryptophan decarboxylase PsiD-like domain-containing protein | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9JFN
 
 | | Arginine decarboxylase in Aspergillus oryzae complexed with agmatine | | Descriptor: | 1,2-ETHANEDIOL, AGMATINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-05 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
1BYB
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYA
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
