1OD5
 
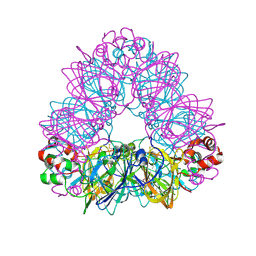 | | Crystal structure of glycinin A3B4 subunit homohexamer | | Descriptor: | CARBONATE ION, GLYCININ, MAGNESIUM ION | | Authors: | Adachi, M, Kanamori, J, Masuda, T, Yagasaki, K, Kitamura, K, Mikami, B, Utsumi, S. | | Deposit date: | 2003-02-13 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Soybean 11S Globulin: Glycinin A3B4 Homohexamer
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Q6D
 
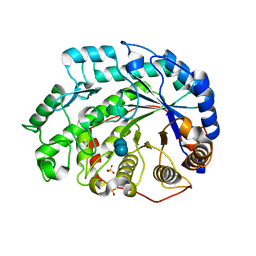 | | Crystal structure of Soybean Beta-Amylase Mutant (M51T) with Increased pH Optimum | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6E
 
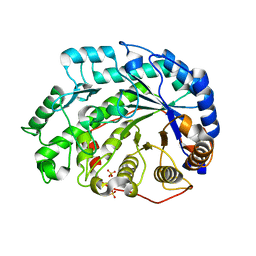 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 5.4 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6G
 
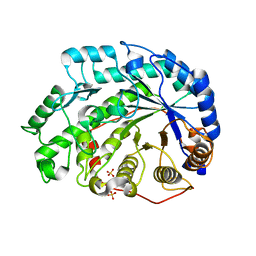 | | Crystal Structure of Soybean Beta-Amylase Mutant (N340T) with Increased pH Optimum | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
4KEP
 
 | | Crystal structure of 4-pyridoxolactonase, wild-type | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, ACETATE ION, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4KEQ
 
 | | Crystal structure of 4-pyridoxolactonase, 5-pyridoxolactone bound | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, 7-hydroxy-6-methylfuro[3,4-c]pyridin-3(1H)-one, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1HV6
 
 | | CRYSTAL STRUCTURE OF ALGINATE LYASE A1-III COMPLEXED WITH TRISACCHARIDE PRODUCT. | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-D-glucopyranuronic acid, ALGINATE LYASE, SULFATE ION | | Authors: | Yoon, H.-J, Hashimoto, W, Miyake, O, Murata, K, Mikami, B. | | Deposit date: | 2001-01-08 | | Release date: | 2001-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
7VGK
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI | | Descriptor: | 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase | | Authors: | Iwase, H, Oiki, S, Mikami, B, Takase, R, Hashimoto, W. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
1NFT
 
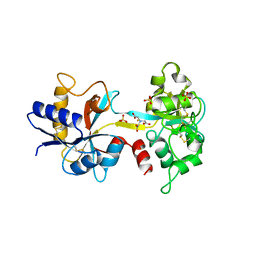 | | OVOTRANSFERRIN, N-TERMINAL LOBE, IRON LOADED OPEN FORM | | Descriptor: | FE (III) ION, NITRILOTRIACETIC ACID, PROTEIN (OVOTRANSFERRIN), ... | | Authors: | Mizutani, K, Yamashita, H, Kurokawa, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1Q1Y
 
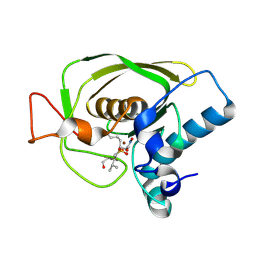 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | Deposit date: | 2003-07-23 | | Release date: | 2004-07-23 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
1NNT
 
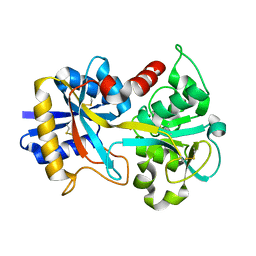 | |
4HA6
 
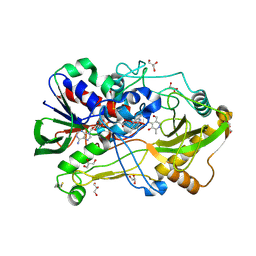 | | Crystal structure of pyridoxine 4-oxidase - pyridoxamine complex | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T. | | Deposit date: | 2012-09-25 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal structure of pyridoxine 4-oxidase from Mesorhizobium loti.
Biochim.Biophys.Acta, 1834, 2013
|
|
4H2Q
 
 | | structure of MHPCO-5HN complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridiine-5-carboxylic acid oxygenase
To be Published
|
|
4H2N
 
 | | Crystal structure of MHPCO, Y270F mutant | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
1J1N
 
 | | Structure Analysis of AlgQ2, A Macromolecule(Alginate)-Binding Periplasmic Protein Of Sphingomonas Sp. A1., Complexed with an Alginate Tetrasaccharide | | Descriptor: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mikami, B, Mishima, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2002-12-11 | | Release date: | 2003-06-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-A resolution
J.BIOL.CHEM., 278, 2003
|
|
7YE3
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
6JBN
 
 | | Crystal structure of Sphingomonas sp. A1 peroxidase EfeB responsible for import of iron | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, OXYGEN MOLECULE, ... | | Authors: | Okumura, K, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rare metal binding by a cell-surface component of bacterial EfeUOB iron importer
To Be Published
|
|
6KJL
 
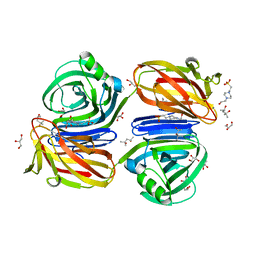 | | Xylanase J from Bacillus sp. strain 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6J33
 
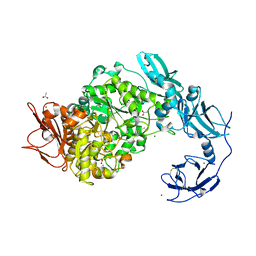 | | Crystal structure of ligand-free of PulA from Klebsiella pneumoniae | | Descriptor: | ACETATE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1Q6C
 
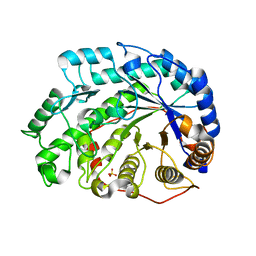 | | Crystal Structure of Soybean Beta-Amylase Complexed with Maltose | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-amylase | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1CQY
 
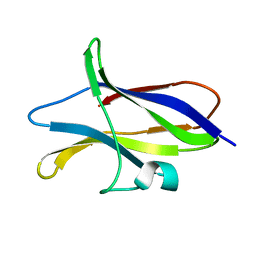 | | STARCH BINDING DOMAIN OF BACILLUS CEREUS BETA-AMYLASE | | Descriptor: | BETA-AMYLASE | | Authors: | Yoon, H.J, Hirata, A, Adachi, M, Sekine, A, Utsumi, S, Mikami, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Separated Starch-Binding Domain of Bacillus cereus B-amylase
To be Published
|
|
3VUP
 
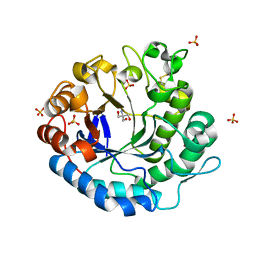 | | Beta-1,4-mannanase from the common sea hare Aplysia kurodai | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase, SULFATE ION | | Authors: | Mizutani, K, Tsuchiya, S, Toyoda, M, Nanbu, Y, Tominaga, K, Yuasa, K, Takahashi, N, Tsuji, A, Mikami, B. | | Deposit date: | 2012-07-04 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of beta-1,4-mannanase from the common sea hare Aplysia kurodai at 1.05 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6J3F
 
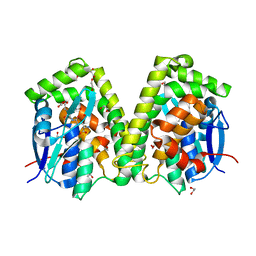 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
7WSS
 
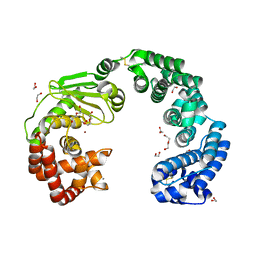 | | Collagenase from Grimontia (Vibrio) hollisae 1706B | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
7XEB
 
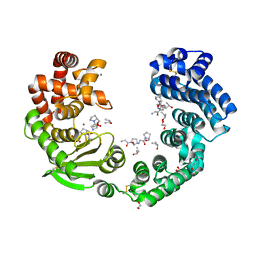 | | Collagenase from Grimontia (Vibrio) hollisae 1706B complexed with Gly-Pro-Hyp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLY-PRO-HYP peptide, ... | | Authors: | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | Deposit date: | 2022-03-30 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
