2OFI
 
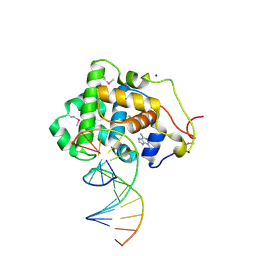 | | Crystal Structure of 3-methyladenine DNA Glycosylase I (TAG) bound to DNA/3mA | | Descriptor: | 3-METHYL-3H-PURIN-6-YLAMINE, 3-methyladenine DNA glycosylase I, constitutive, ... | | Authors: | Metz, A.H, Hollis, T, Eichman, B.F. | | Deposit date: | 2007-01-03 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG).
Embo J., 26, 2007
|
|
2OFK
 
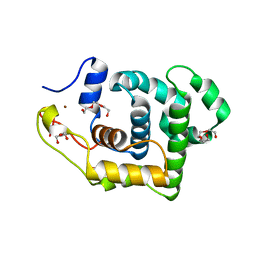 | | Crystal Structure of 3-methyladenine DNA glycosylase I (TAG) | | Descriptor: | 3-methyladenine DNA glycosylase I, constitutive, TRIETHYLENE GLYCOL, ... | | Authors: | Metz, A.H, Hollis, T, Eichman, B.F. | | Deposit date: | 2007-01-03 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG).
Embo J., 26, 2007
|
|
5OG7
 
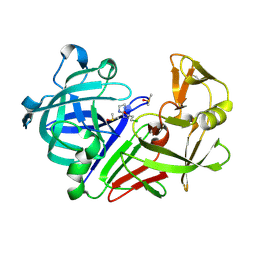 | | Endothiapepsin in complex with hydrazide fragment | | Descriptor: | 3-chloranyl-~{N}'-(4,5-dihydro-1~{H}-imidazol-3-ium-2-yl)propanehydrazide, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Metz, A, Klauser, P.C, Heine, A, Klebe, G. | | Deposit date: | 2017-07-12 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Endothiapepsin in complex with hydrazide fragment
To Be Published
|
|
1SCN
 
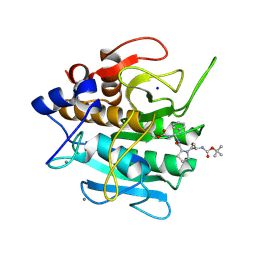 | | INACTIVATION OF SUBTILISIN CARLSBERG BY N-(TERT-BUTOXYCARBONYL-ALANYL-PROLYL-PHENYLALANYL)-O-BENZOL HYDROXYLAMINE: FORMATION OF COVALENT ENZYME-INHIBITOR LINKAGE IN THE FORM OF A CARBAMATE DERIVATIVE | | Descriptor: | CALCIUM ION, N-(tert-butoxycarbonyl)-L-alanyl-N-[(1R)-1-(carboxyamino)-2-phenylethyl]-L-prolinamide, SODIUM ION, ... | | Authors: | Steinmetz, A.C.U, Demuth, H.-U, Ringe, D. | | Deposit date: | 1994-03-02 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inactivation of subtilisin Carlsberg by N-((tert-butoxycarbonyl)alanylprolylphenylalanyl)-O-benzolhydroxyl- amine: formation of a covalent enzyme-inhibitor linkage in the form of a carbamate derivative.
Biochemistry, 33, 1994
|
|
3DJL
 
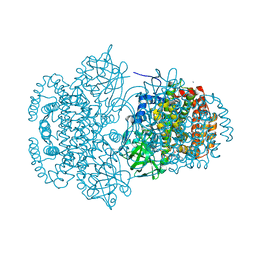 | |
7GRZ
 
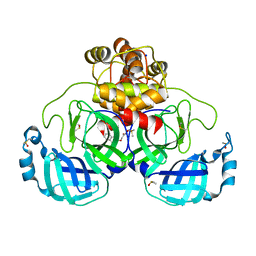 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-22 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N,N-dimethyl-2-[(naphthalen-2-yl)oxy]acetamide, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS2
 
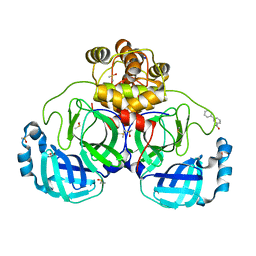 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-25 | | Descriptor: | 3-(pyridin-3-yl)benzoic acid, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS6
 
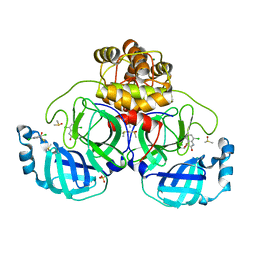 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-29 | | Descriptor: | (3S)-4,7-dichloro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS3
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-26 | | Descriptor: | (6-phenylpyridin-3-yl)methanamine, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-28 | | Descriptor: | (3R)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRG
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-3 | | Descriptor: | 3,5-dichloropyridin-4-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRT
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-16 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2,3-dihydro-1-benzofuran-5-yl)methyl]benzamide, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRR
 
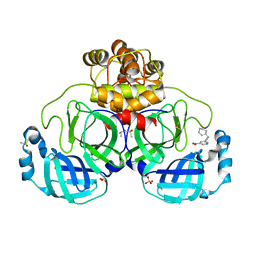 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-14 | | Descriptor: | 3C-like proteinase nsp5, 5-(3-cyclohexylprop-1-yn-1-yl)pyridine-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRY
 
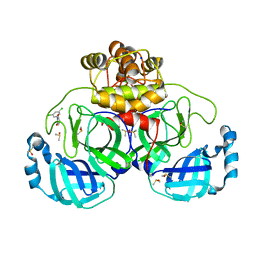 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-21 | | Descriptor: | 1-(3,5-dichlorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRH
 
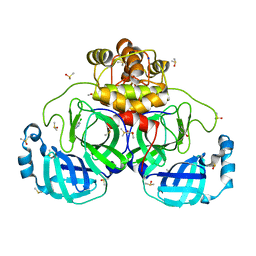 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-4 | | Descriptor: | 3C-like proteinase nsp5, 5-chloropyridin-3-ol, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRP
 
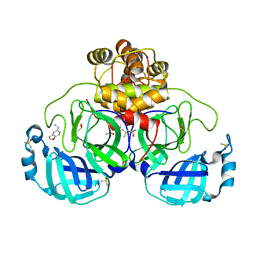 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-12 | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)methanamine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-7 | | Descriptor: | (6-fluoro-2H,4H-1,3-benzodioxin-8-yl)methanol, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRI
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-5 | | Descriptor: | (1S)-1-(1H-pyrazol-5-yl)ethan-1-ol, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS4
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-27 | | Descriptor: | 3C-like proteinase nsp5, 7-(hydroxymethyl)-3-methyl-6~{H}-[1,3]thiazolo[3,2-a]pyrimidin-5-one, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRF
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-2 | | Descriptor: | 3C-like proteinase nsp5, 5-bromopyridin-3-amine, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRL
 
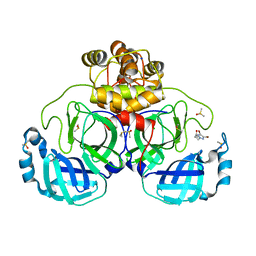 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-8 | | Descriptor: | 3C-like proteinase nsp5, 4-(4,5-dibromo-2H-1,2,3-triazol-2-yl)butan-2-one, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRO
 
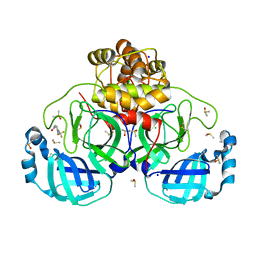 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-11 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRQ
 
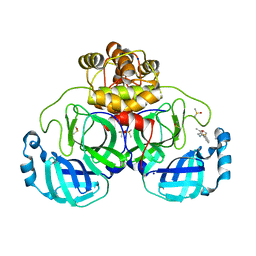 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-13 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRN
 
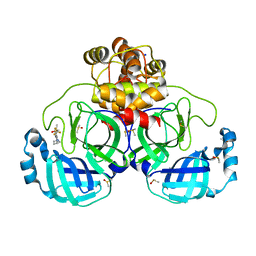 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-10 | | Descriptor: | 2-[(3S)-pyrrolidin-3-yl]-5-(trifluoromethyl)-1H-benzimidazole, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-6 | | Descriptor: | (5-chloro-1-benzothiophen-3-yl)methanol, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
