3T38
 
 | | Corynebacterium glutamicum thioredoxin-dependent arsenate reductase Cg_ArsC1' | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Arsenate Reductase | | Authors: | Messens, J, Wahni, K, Dufe, T.V. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-23 | | Last modified: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Corynebacterium glutamicum survives arsenic stress with arsenate reductases coupled to two distinct redox mechanisms.
Mol.Microbiol., 82, 2011
|
|
4FIW
 
 | |
3RH0
 
 | |
1RXE
 
 | | ArsC complexed with MNB | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Arsenate reductase, PERCHLORATE ION, ... | | Authors: | Messens, J, Van Molle, I, Vanhaesebrouck, P, Limbourg, M, Van Belle, K, Wahni, K, Martins, J.C, Loris, R, Wyns, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a triple mutant of pI258 arsenate reductase from Staphylococcus aureus and its 5-thio-2-nitrobenzoic acid adduct.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RXI
 
 | | pI258 arsenate reductase (ArsC) triple mutant C10S/C15A/C82S | | Descriptor: | Arsenate reductase, CHLORIDE ION, PERCHLORATE ION, ... | | Authors: | Messens, J, Van Molle, I, Vanhaesebrouck, P, Limbourg, M, Van Belle, K, Wahni, K, Martins, J.C, Loris, R, Wyns, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a triple mutant of pI258 arsenate reductase from Staphylococcus aureus and its 5-thio-2-nitrobenzoic acid adduct.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1LK0
 
 | | Disulfide intermediate of C89L Arsenate reductase from pI258 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, arsenate reductase | | Authors: | Messens, J, Martins, J.C, Van Belle, K, Brosens, E, Desmyter, A, De Gieter, M, Wieruszeski, J.M, Willem, R, Wyns, L, Zegers, I. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LJL
 
 | | Wild Type pI258 S. aureus arsenate reductase | | Descriptor: | POTASSIUM ION, arsenate reductase | | Authors: | Messens, J, Martins, J.C, Van Belle, K, Brosens, E, Desmyter, A, De Gieter, M, Wieruszeski, J.M, Willem, R, Wyns, L, Zegers, I. | | Deposit date: | 2002-04-21 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2PQX
 
 | | E. coli RNase 1 (in vivo folded) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Ribonuclease I | | Authors: | Messens, J, Loris, R, Wyns, L. | | Deposit date: | 2007-05-03 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of E. coli ribonuclease 1
To be Published
|
|
2PQY
 
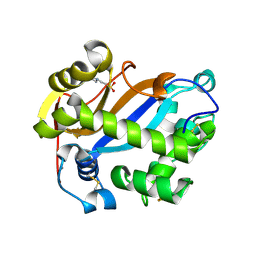 | | E. coli RNase 1 (in vitro refolded with DsbA only) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Ribonuclease I | | Authors: | Messens, J, Loris, R. | | Deposit date: | 2007-05-03 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The oxidase DsbA folds a protein with a nonconsecutive disulfide.
J.Biol.Chem., 282, 2007
|
|
1QOS
 
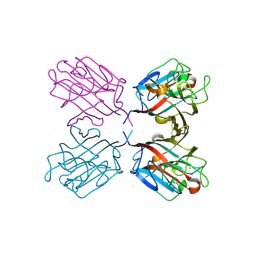 | | lectin UEA-II complexed with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-16 | | Release date: | 2000-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
8PR6
 
 | |
6TR8
 
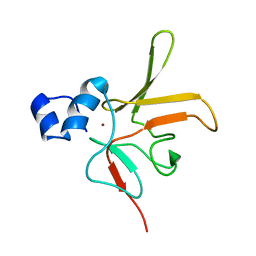 | | Corynebacterium diphtheriae methionine sulfoxide reductase B (MsrB) solution structure - reduced form | | Descriptor: | Peptide-methionine (R)-S-oxide reductase, ZINC ION | | Authors: | Volkov, A.N, Tossounian, M.A, Buts, L, Messens, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Methionine sulfoxide reductase B fromCorynebacterium diphtheriaecatalyzes sulfoxide reduction via an intramolecular disulfide cascade.
J.Biol.Chem., 295, 2020
|
|
6EP6
 
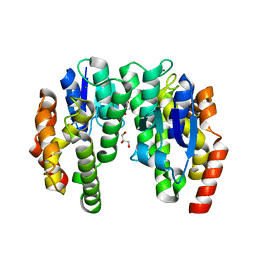 | | ARABIDOPSIS THALIANA GSTU23, reduced | | Descriptor: | GLYCEROL, Glutathione S-transferase U23, MAGNESIUM ION | | Authors: | Tossounian, M.A, Van Molle, I, Wahni, K, Jacques, S, Vertommen, D, Gevaert, K, Van Breusegem, F, Young, D, Rosado, L, Messens, J. | | Deposit date: | 2017-10-10 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
6ZUI
 
 | | Crystal structure of the Cys-Ser mutant of the cpYFP-based biosensor for hypochlorous acid | | Descriptor: | HTH-type transcriptional repressor NemR,Green fluorescent protein,Green fluorescent protein,HTH-type transcriptional repressor NemR | | Authors: | Tossounian, M.A, Van Molle, I, Messens, J. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.200082 Å) | | Cite: | Hypocrates is a genetically encoded fluorescent biosensor for (pseudo)hypohalous acids and their derivatives.
Nat Commun, 13, 2022
|
|
6QFS
 
 | | Chargeless variant of the Cellulose-binding domain from Cellulomonas fimi | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ... | | Authors: | Young, D.R, Hoejgaard, C, Messens, J, Winther, J.R. | | Deposit date: | 2019-01-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Charge Interactions in a Highly Charge-depleted Protein
J.Am.Chem.Soc., 2021
|
|
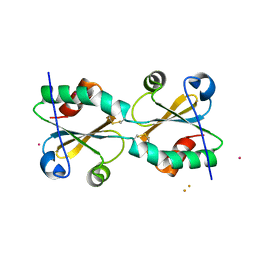
3DIE
 
 | |
4D7L
 
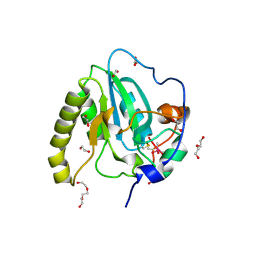 | | Methionine sulfoxide reductase A of Corynebacterium diphtheriae | | Descriptor: | CACODYLATE ION, PEPTIDE METHIONINE SULFOXIDE REDUCTASE MSRA, SULFATE ION, ... | | Authors: | Van Molle, I, Tossounian, M.A, Pedre, B, Wahni, K, Vertommen, D, Messens, J. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Corynebacterium Diphtheriae Methionine Sulfoxide Reductase a Exploits a Unique Mycothiol Redox Relay Mechanism.
J.Biol.Chem., 290, 2015
|
|
2CD7
 
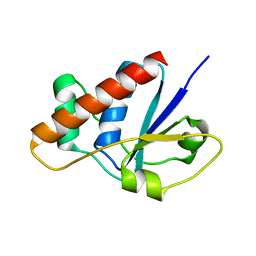 | | Staphylococcus aureus pI258 arsenate reductase (ArsC) H62Q mutant | | Descriptor: | PROTEIN ARSC, SODIUM ION | | Authors: | Buts, L, Roos, G, Van Belle, K, Brosens, E, Loris, R, Wyns, L, Messens, J. | | Deposit date: | 2006-01-23 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interplay between Ion Binding and Catalysis in the Thioredoxin-Coupled Arsenate Reductase Family.
J.Mol.Biol., 360, 2006
|
|
5C04
 
 | |
4N30
 
 | |
2Z70
 
 | | E.coli RNase 1 in complex with d(CGCGATCGCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DTP*DCP*DGP*DCP*DG)-3'), Ribonuclease I | | Authors: | Martinez-Rodriguez, S, Loris, R, Messens, J. | | Deposit date: | 2007-08-09 | | Release date: | 2008-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonspecific base recognition mediated by water bridges and hydrophobic stacking in ribonuclease I from Escherichia coli
Protein Sci., 17, 2008
|
|
2FXI
 
 | |
5G1K
 
 | | A triple mutant of DsbG engineered for denitrosylation | | Descriptor: | SULFATE ION, THIOL DISULFIDE INTERCHANGE PROTEIN DSBG | | Authors: | Tamu Dufe, V, Van Molle, I, Lafaye, C, Wahni, K, Boudier, A, Leroy, P, Collet, J.F, Messens, J. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Sulfur Denitrosylation by an Engineered Trx-Like Dsbg Enzyme Identifies Nucleophilic Cysteine Hydrogen Bonds as Key Functional Determinant.
J.Biol.Chem., 291, 2016
|
|
1DZQ
 
 | | LECTIN UEA-II COMPLEXED WITH GALACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN II, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 2000-03-07 | | Release date: | 2000-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
5G1L
 
 | | A double mutant of DsbG engineered for denitrosylation | | Descriptor: | SULFATE ION, THIOL DISULFIDE INTERCHANGE PROTEIN DSBG | | Authors: | Tamu Dufe, V, Van Molle, I, Lafaye, C, Wahni, K, Boudier, A, Leroy, P, Collet, J.F, Messens, J. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfur Denitrosylation by an Engineered Trx-Like Dsbg Enzyme Identifies Nucleophilic Cysteine Hydrogen Bonds as Key Functional Determinant.
J.Biol.Chem., 291, 2016
|
|
