4W7N
 
 | |
8OY1
 
 | |
2YMW
 
 | |
1AZW
 
 | | PROLINE IMINOPEPTIDASE FROM XANTHOMONAS CAMPESTRIS PV. CITRI | | Descriptor: | PROLINE IMINOPEPTIDASE | | Authors: | Medrano, F.J, Alonso, J, Garcia, J.L, Romero, A, Bode, W, Gomis-Ruth, F.X. | | Deposit date: | 1997-11-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of proline iminopeptidase from Xanthomonas campestris pv. citri: a prototype for the prolyl oligopeptidase family.
EMBO J., 17, 1998
|
|
4W7O
 
 | |
4W7L
 
 | |
4W7J
 
 | |
4W7M
 
 | |
4W7K
 
 | |
8PAQ
 
 | |
5ABQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF FUNGAL VERSATILE PEROXIDASE FROM PLEUROTUS ERYNGII. MUTANT VPi-SS. MUTATED RESIDUES T2K, A49C, A61C, D69S, T70D, S86E, A131K, D146T, Q202L, Q219K, H232E, Q239R, L288R, S301K, A308R,A309K AND A314R. | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2015-08-07 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Improving the Ph-Stability of Versatile Peroxidase by Comparative Structural Analysis with a Naturally-Stable Manganese Peroxidase.
Plos One, 10, 2015
|
|
5ABN
 
 | | CRYSTAL STRUCTURE ANALYSIS OF FUNGAL VERSATILE PEROXIDASE FROM PLEUROTUS ERYNGII. MUTANT VPi. MUTATED RESIDUES D69S, T70D, S86E, D146T, Q202L, H232E, Q239R AND S301K. | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2015-08-07 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Improving the Ph-Stability of Versatile Peroxidase by Comparative Structural Analysis with a Naturally-Stable Manganese Peroxidase.
Plos One, 10, 2015
|
|
5ABO
 
 | | CRYSTAL STRUCTURE ANALYSIS OF FUNGAL VERSATILE PEROXIDASE FROM PLEUROTUS ERYNGII. MUTANT VPi-br. MUTATED RESIDUES T2K, D69S, T70D, S86E, A131K, D146T, Q202L, Q219K, H232E, Q239R, L288R, S301K, A308R, A309K AND A314R. | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE VPL2 | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2015-08-07 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Improving the Ph-Stability of Versatile Peroxidase by Comparative Structural Analysis with a Naturally-Stable Manganese Peroxidase.
Plos One, 10, 2015
|
|
4BM2
 
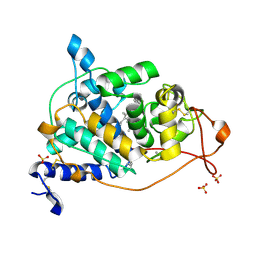 | | CRYSTAL STRUCTURE OF MANGANESE PEROXIDASE 4 FROM PLEUROTUS OSTREATUS - CRYSTAL FORM II | | Descriptor: | CALCIUM ION, MANGANESE PEROXIDASE 4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
4BM0
 
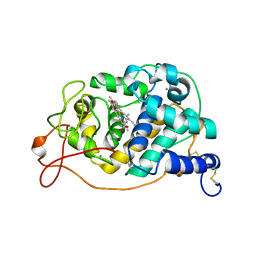 | | CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM VII | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE I | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
4BM3
 
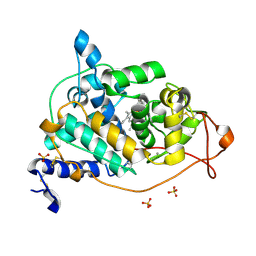 | | CRYSTAL STRUCTURE OF MANGANESE PEROXIDASE 4 FROM PLEUROTUS OSTREATUS - CRYSTAL FORM III | | Descriptor: | CALCIUM ION, MANGANESE PEROXIDASE 4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
4BLX
 
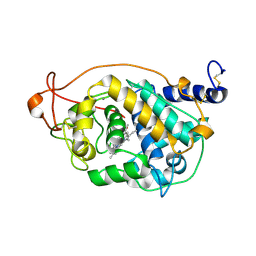 | | CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM IV | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE I | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
4BLZ
 
 | | CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE I FROM PLEUROTUS OSTREATUS - CRYSTAL FORM VI | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, VERSATILE PEROXIDASE I | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
4BM4
 
 | | CRYSTAL STRUCTURE OF MANGANESE PEROXIDASE 4 FROM PLEUROTUS OSTREATUS - CRYSTAL FORM IV | | Descriptor: | CALCIUM ION, MANGANESE PEROXIDASE 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
3UOR
 
 | |
1AU8
 
 | | HUMAN CATHEPSIN G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-[(1R)-5-amino-1-phosphonopentyl]-L-prolinamide | | Authors: | Medrano, F.J, Bode, W, Banbula, A, Potempa, J. | | Deposit date: | 1997-09-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HUMAN CATHEPSIN G
to be published
|
|
5ANH
 
 | |
5FNB
 
 | | CRYSTAL STRUCTURE OF FUNGAL VERSATILE PEROXIDASE FROM PLEUROTUS ERYNGII SEPTUPLE MUTANT E37K, H39R, V160A, T184M, Q202L, D213A & G330R | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Unveiling the Basis of Alkaline Stability of an Evolved Versatile Peroxidase.
Biochem.J., 473, 2016
|
|
5FNE
 
 | |
4CZN
 
 | |
