1KK1
 
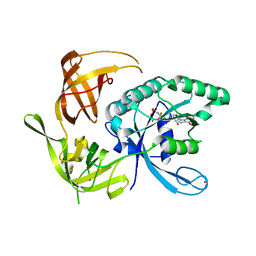 | | Structure of the large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi-G235D mutant complexed with GDPNP-Mg2+ | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
4ZGQ
 
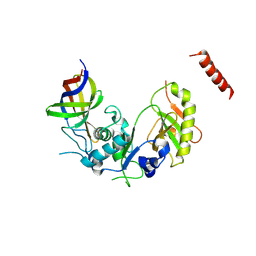 | | Structure of Cdc123 bound to eIF2-gammaDIII domain | | Descriptor: | Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
4ZGN
 
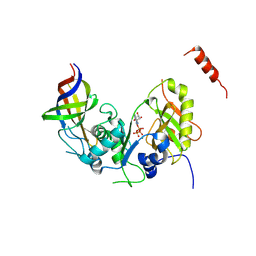 | | Structure Cdc123 complexed with the C-terminal domain of eIF2gamma | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
1ZQ1
 
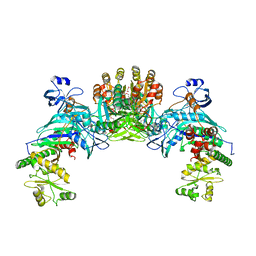 | | Structure of GatDE tRNA-Dependent Amidotransferase from Pyrococcus abyssi | | Descriptor: | ASPARTIC ACID, Glutamyl-tRNA(Gln) amidotransferase subunit D, Glutamyl-tRNA(Gln) amidotransferase subunit E | | Authors: | Schmitt, E, Panvert, M, Blanquet, S, Mechulam, Y. | | Deposit date: | 2005-05-18 | | Release date: | 2005-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for tRNA-Dependent Amidotransferase Function
Structure, 13, 2005
|
|
4ZGO
 
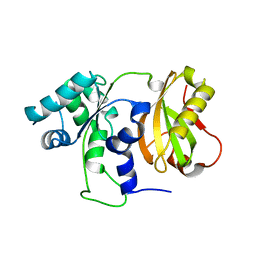 | | Structure of C-terminally truncated Cdc123 from Schizosaccharomyces pombe | | Descriptor: | Cell division cycle protein 123 | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
4ZGP
 
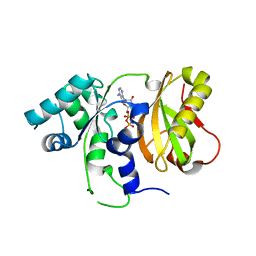 | | Structure of Cdc123 from Schizosaccharomyces pombe | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division cycle protein 123 | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
1MKH
 
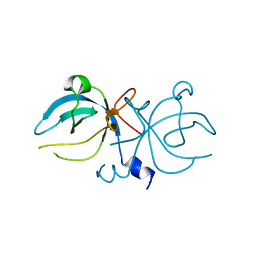 | |
1YZ6
 
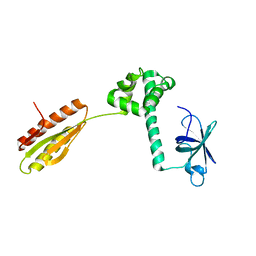 | |
1PFY
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH METHIONYL SULPHAMOYL ADENOSINE | | Descriptor: | 5'-O-[(L-METHIONYL)-SULPHAMOYL]ADENOSINE, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PFU
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH METHIONINE PHOSPHINATE | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PG0
 
 | | Methionyl-trna synthetase from escherichia coli complexed with methioninyl adenylate | | Descriptor: | L-METHIONYL ADENYLATE, Methionyl-tRNA synthetase | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PFW
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH TRIFLUOROMETHIONINE | | Descriptor: | 2-AMINO-4-TRIFLUOROMETHYLSULFANYL-BUTYRIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PFV
 
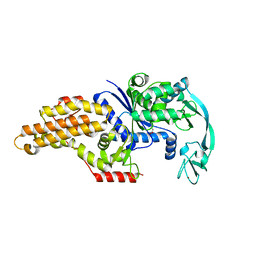 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH DIFLUOROMETHIONINE | | Descriptor: | Methionyl-tRNA synthetase, S-(DIFLUOROMETHYL)HOMOCYSTEINE, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PG2
 
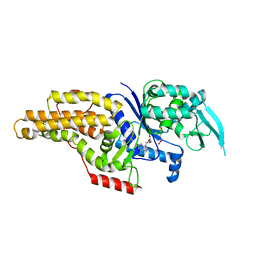 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH METHIONINE AND ADENOSINE | | Descriptor: | ADENOSINE, METHIONINE, Methionyl-tRNA synthetase | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
6SPR
 
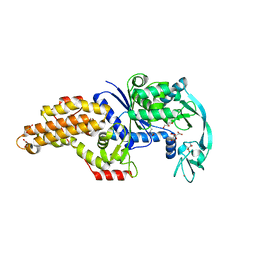 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SPP
 
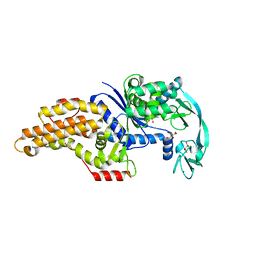 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 | | Descriptor: | CITRIC ACID, GLYCEROL, Methionine--tRNA ligase, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SWD
 
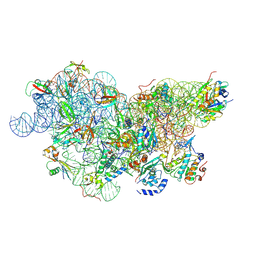 | | IC2 body model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6SWC
 
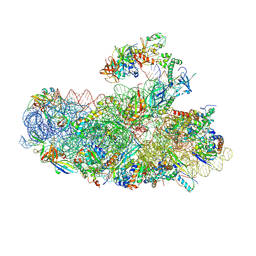 | | IC2B model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6SWE
 
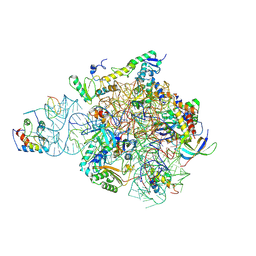 | | IC2 head of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6SPO
 
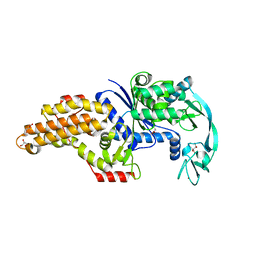 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SPQ
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SW9
 
 | | IC2A model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6SPN
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
3FRH
 
 | | Structure of the 16S rRNA methylase RmtB, P21 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3FZG
 
 | | Structure of the 16S rRNA methylase ArmA | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
