4AD8
 
 | | Crystal structure of a deletion mutant of Deinococcus radiodurans RecN | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.998 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4ABX
 
 | | Crystal structure of Deinococcus radiodurans RecN coiled-coil domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
2JG1
 
 | | STRUCTURE OF Staphylococcus aureus D-TAGATOSE-6-PHOSPHATE KINASE with cofactor and substrate | | Descriptor: | 6-O-phosphono-beta-D-tagatofuranose, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Miallau, L, Hunter, W.N, McSweeney, S.M, Leonard, G.A. | | Deposit date: | 2007-02-07 | | Release date: | 2007-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Staphylococcus Aureus D-Tagatose-6-Phosphate Kinase Implicate Domain Motions in Specificity and Mechanism.
J.Biol.Chem., 282, 2007
|
|
2JGV
 
 | | STRUCTURE OF Staphylococcus aureus D-TAGATOSE-6-PHOSPHATE KINASE in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TAGATOSE-6-PHOSPHATE KINASE | | Authors: | Miallau, L, Hunter, W.N, McSweeney, S.M, Leonard, G.A. | | Deposit date: | 2007-02-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Staphylococcus Aureus D-Tagatose-6-Phosphate Kinase Implicate Domain Motions in Specificity and Mechanism.
J.Biol.Chem., 282, 2007
|
|
1HJ8
 
 | | 1.00 AA Trypsin from Atlantic Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Leiros, H.-K.S, Mcsweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HJ9
 
 | | Atomic resolution structures of trypsin provide insight into structural radiation damage | | Descriptor: | ANILINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, McSweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-10 | | Release date: | 2002-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HZZ
 
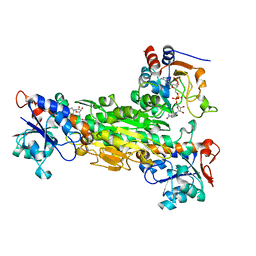 | | THE ASYMMETRIC COMPLEX OF THE TWO NUCLEOTIDE-BINDING COMPONENTS (DI, DIII) OF PROTON-TRANSLOCATING TRANSHYDROGENASE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTON-TRANSLOCATING NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE SUBUNIT PNTAA, ... | | Authors: | Cotton, N.P.J, White, S.A, Peake, S.J, McSweeney, S, Jackson, J.B. | | Deposit date: | 2001-01-27 | | Release date: | 2001-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an asymmetric complex of the two nucleotide binding components of proton-translocating transhydrogenase.
Structure, 9, 2001
|
|
1I4U
 
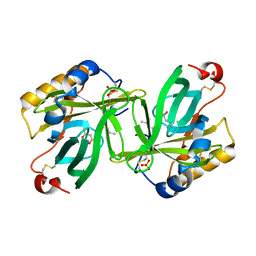 | | THE C1 SUBUNIT OF ALPHA-CRUSTACYANIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CRUSTACYANIN, SULFATE ION | | Authors: | Gordon, E.J, Leonard, G.A, McSweeney, S, Zagalsky, P.F. | | Deposit date: | 2001-02-23 | | Release date: | 2001-09-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The C1 subunit of alpha-crustacyanin: the de novo phasing of the crystal structure of a 40 kDa homodimeric protein using the anomalous scattering from S atoms combined with direct methods.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4B7W
 
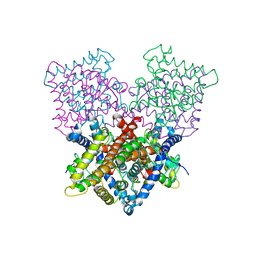 | | Ligand binding domain human hepatocyte nuclear factor 4alpha: Apo form | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 4-ALPHA | | Authors: | Dudasova, Z, Okvist, M, Kretova, M, Ondrovicova, G, Skrabana, R, LeGuevel, R, Salbert, G, Leonard, G, McSweeney, S, Barath, P. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Fatty Acids are not Essential Structural Components of Hepatocyte Nuclear Factor 4Alpha
To be Published
|
|
3I34
 
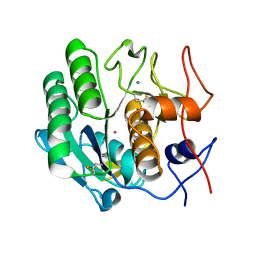 | | Proteinase K by LB Nanotemplate Method after high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, MERCURY (II) ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Radiation damage study of Proteinase K at ID14-2 beamline at ESRF
To be Published
|
|
3I6J
 
 | | Ribonuclease A by Classical hanging drop method after high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3I2Y
 
 | | Proteinase K by Classical hanging drop Method before high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.995 Å) | | Cite: | Atomic structure and radiation resistance of Langmuir-Blodgett protein crystals
To be Published
|
|
3I30
 
 | | Proteinase K by Classical hanging drop Method after high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.992 Å) | | Cite: | Atomic structure and radiation resistance of Langmuir-Blodgett protein crystals
To be Published
|
|
3I6F
 
 | | Ribonuclease A by Classical hanging drop method before high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3I67
 
 | | Ribonuclease A by LB nanotemplate method after high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3I37
 
 | | Proteinase K by LB Nanotemplate Method before high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.995 Å) | | Cite: | Atomic structure and radiation resistance of Langmuir-Blodgett protein crystals
To Be Published
|
|
3I6H
 
 | | Ribonuclease A by LB nanotemplate method before high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
7MHF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHL
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHM
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
2YEU
 
 | | Structural and functional insights of DR2231 protein, the MazG-like nucleoside triphosphate pyrophosphohydrolase from Deinococcus radiodurans, complex with Gd | | Descriptor: | DR2231, GADOLINIUM ATOM, GLYCEROL, ... | | Authors: | Goncalves, A.M.D, de Sanctis, D, McSweeney, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2011-07-06 | | Last modified: | 2011-09-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
2YF3
 
 | | Crystal structure of DR2231, the MazG-like protein from Deinococcus radiodurans, complex with manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE, ... | | Authors: | Goncalves, A.M.D, deSanctis, D, McSweeney, S.M. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
2YF4
 
 | | Crystal structure of DR2231, the MazG-like protein from Deinococcus radiodurans, Apo structure | | Descriptor: | GLYCEROL, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE, SULFATE ION | | Authors: | Goncalves, A.M.D, deSanctis, D, McSweeney, S.M. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
2YFC
 
 | | STRUCTURAL AND FUNCTIONAL INSIGHTS OF DR2231 PROTEIN, THE MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE FROM DEINOCOCCUS RADIODURANS, COMPLEXED WITH Mn and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Goncalves, A.M.D, De Sanctis, D, Mcsweeney, S.M. | | Deposit date: | 2011-04-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
