7OSB
 
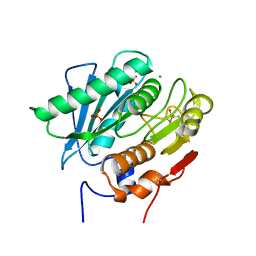 | | Crystal Structure of a Double Mutant PETase (S238F/W159H) from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Shakespeare, T.J, Zahn, M, Allen, M.D, McGeehan, J.E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation.
ChemSusChem, 15, 2022
|
|
7ON9
 
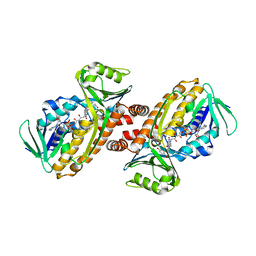 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Zahn, M, McGeehan, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
5OMU
 
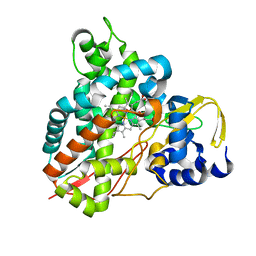 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMR
 
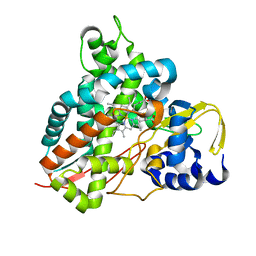 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with vanillin. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, GcoA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
3FYA
 
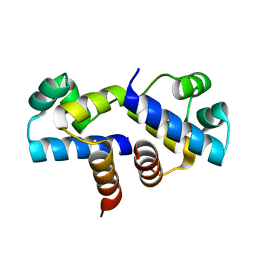 | | Crystal Structure of an R35A mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5OMS
 
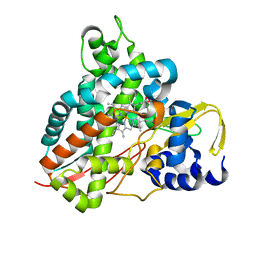 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaethol. | | Descriptor: | 2-ethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
3G5G
 
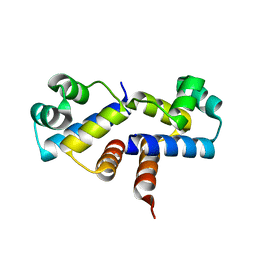 | | Crystal Structure of the Wild-Type Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5OGX
 
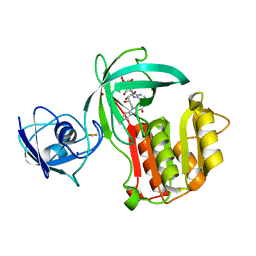 | | Crystal structure of Amycolatopsis cytochrome P450 reductase GcoB. | | Descriptor: | BROMIDE ION, Cytochrome P450 reductase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5NCB
 
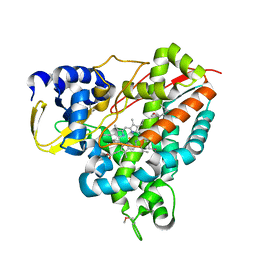 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaiacol. | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-07-04 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
3UFD
 
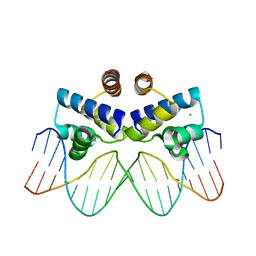 | | C.Esp1396I bound to its highest affinity operator site OM | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*TP*GP*TP*AP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*TP*AP*CP*A)-3'), ... | | Authors: | Ball, N.J, McGeehan, J.E, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2011-11-01 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of differential DNA sequence recognition by restriction-modification controller proteins.
Nucleic Acids Res., 40, 2012
|
|
8AYV
 
 | |
7QJR
 
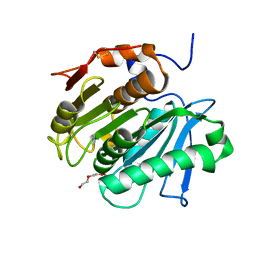 | | Crystal structure of cutinase 1 from Thermobifida fusca DSM44342 (703) | | Descriptor: | Cutinase 1, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJT
 
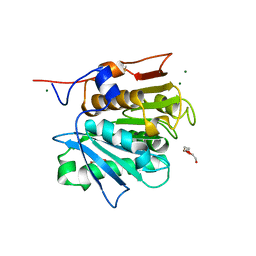 | | Crystal structure of a cutinase enzyme from Thermobifida cellulosilytica TB100 (711) | | Descriptor: | GLYCEROL, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJQ
 
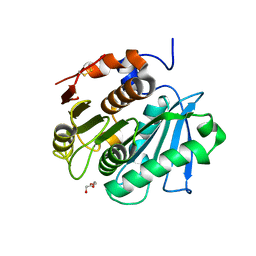 | | Crystal structure of a cutinase enzyme from Thermobifida fusca NTU22 (702) | | Descriptor: | Acetylxylan esterase, DI(HYDROXYETHYL)ETHER | | Authors: | Zahn, M, Gill, R.S, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJP
 
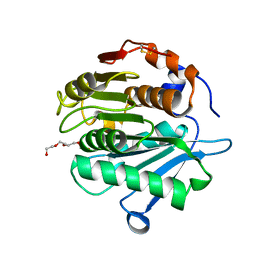 | | Crystal structure of a cutinase enzyme from Saccharopolyspora flava (611) | | Descriptor: | Cutinase, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJO
 
 | |
7QJM
 
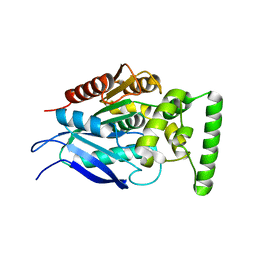 | |
7QJN
 
 | | Crystal structure of an alpha/beta-hydrolase enzyme from Candidatus Kryptobacter tengchongensis (306) | | Descriptor: | Dienelactone hydrolase, PHOSPHATE ION | | Authors: | Zahn, M, Gill, R.S, Erickson, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJS
 
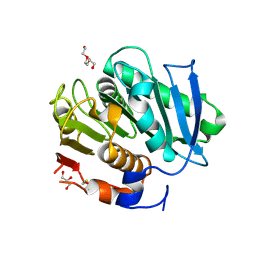 | | Crystal structure of a cutinase enzyme from Thermobifida fusca YX (705) | | Descriptor: | Cutinase 2, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
6HQM
 
 | | Crystal structure of GcoA F169I bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQP
 
 | | Crystal structure of GcoA F169V bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQS
 
 | | Crystal structure of GcoA F169S bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQK
 
 | | Crystal structure of GcoA F169A bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQR
 
 | | Crystal structure of GcoA F169H bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQN
 
 | | Crystal structure of GcoA F169L bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
