3CLC
 
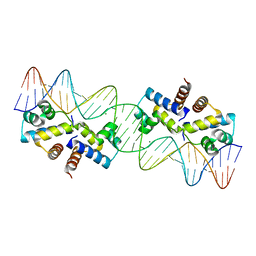 | | Crystal Structure of the Restriction-Modification Controller Protein C.Esp1396I Tetramer in Complex with its Natural 35 Base-Pair Operator | | Descriptor: | 35-MER, MAGNESIUM ION, Regulatory protein | | Authors: | McGeehan, J.E, Streeter, S.D, Thresh, S.J, Ball, N, Ravelli, R.B, Kneale, G.G. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the genetic switch that regulates the expression of restriction-modification genes.
Nucleic Acids Res., 36, 2008
|
|
4GWA
 
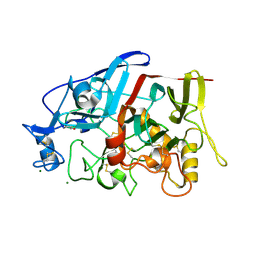 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1Y7Y
 
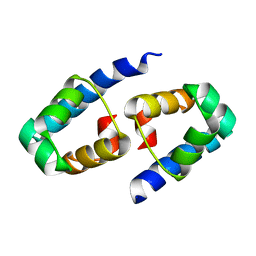 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | Descriptor: | C.AhdI | | Authors: | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
3LFP
 
 | |
3LIS
 
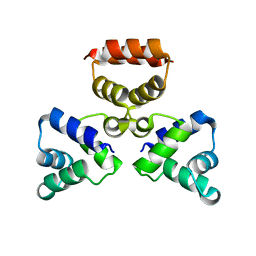 | |
3S8Q
 
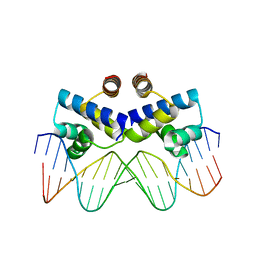 | | Crystal structure of the R-M controller protein C.Esp1396I OL operator complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*AP*CP*TP*TP*AP*TP*AP*GP*TP*CP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*GP*AP*CP*TP*AP*TP*AP*AP*GP*TP*CP*AP*CP*A)-3'), R-M CONTROLLER PROTEIN | | Authors: | McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2011-05-30 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex.
Nucleic Acids Res., 40, 2012
|
|
4IPM
 
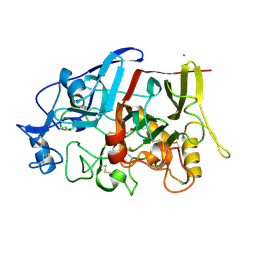 | | Crystal structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with thiocellobiose | | Descriptor: | ACETATE ION, CALCIUM ION, GH7 family protein, ... | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8AIS
 
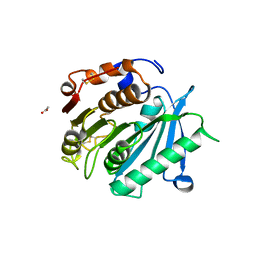 | | Crystal structure of cutinase PsCut from Pseudomonas saudimassiliensis | | Descriptor: | ACETATE ION, Lipase 1 | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AIR
 
 | | Crystal structure of cutinase RgCutII from Rhizobacter gummiphilus | | Descriptor: | ACETATE ION, RgCutII | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
4HAQ
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4X4E
 
 | |
4X4I
 
 | |
4X4G
 
 | |
4X4F
 
 | |
7Q06
 
 | | Crystal structure of TPADO in complex with 2-OH-TPA | | Descriptor: | 2-Hydroxyterephthalic acid, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q05
 
 | | Crystal structure of TPADO in complex with TPA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q2A
 
 | | Crystal structure of AphC in complex with 4-ethylcatechol | | Descriptor: | 4-ethylbenzene-1,2-diol, CALCIUM ION, Catechol 2,3-dioxygenase, ... | | Authors: | Zahn, M, Grigg, J.C, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of a phylogenetically distinct extradiol dioxygenase involved in the bacterial catabolism of lignin-derived aromatic compounds.
J.Biol.Chem., 298, 2022
|
|
7Q04
 
 | | Crystal structure of TPADO in a substrate-free state | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QWT
 
 | | Rieske non-heme iron monooxygenase for guaiacol O-demethylation | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Rieske (2Fe-2S) domain protein | | Authors: | Hinchen, D.J, Zahn, M, Bleem, A, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Discovery, characterization, and metabolic engineering of Rieske non-heme iron monooxygenases for guaiacol O-demethylation
Chem Catal, 2022
|
|
8ABV
 
 | | Crystal structure of SpLdpA in complex with erythro-DGPD | | Descriptor: | (1R,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, GLYCEROL, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABT
 
 | | Crystal structure of NaLdpA in complex with the product analog Resveratrol | | Descriptor: | RESVERATROL, SULFATE ION, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABU
 
 | | Crystal structure of NaLdpA mutant H97Q in complex with erythro-DGPD | | Descriptor: | (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABW
 
 | | Crystal structure of SpLdpA in complex with threo-DGPD | | Descriptor: | (1R,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SULFATE ION, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8AIT
 
 | | Crystal structure of cutinase PbauzCut from Pseudomonas bauzanensis | | Descriptor: | Cutinase, SULFATE ION | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
7OSB
 
 | | Crystal Structure of a Double Mutant PETase (S238F/W159H) from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Shakespeare, T.J, Zahn, M, Allen, M.D, McGeehan, J.E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation.
ChemSusChem, 15, 2022
|
|
