1N1S
 
 | | Trypanosoma rangeli sialidase | | Descriptor: | SULFATE ION, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-20 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The high resolution structures of free and
inhibitor-bound Trypanosoma rangeli
sialidase and its comparison with T. cruzi
trans-sialidase
J.Mol.Biol., 325, 2003
|
|
3H8Z
 
 | | The Crystal Structure of the Tudor Domains from FXR2 | | Descriptor: | Fragile X mental retardation syndrome-related protein 2 | | Authors: | Amaya, M.F, Dong, A, Adams-Cioaba, M.A, Guo, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2.
Plos One, 5, 2010
|
|
3I91
 
 | | Crystal structure of human chromobox homolog 8 (CBX8) with H3K9 peptide | | Descriptor: | Chromobox protein homolog 8, H3K9 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
3FDT
 
 | | Crystal structure of the complex of human chromobox homolog 5 (CBX5) with H3K9(me)3 peptide | | Descriptor: | Chromobox protein homolog 5, H3K9(me)3 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
1N1T
 
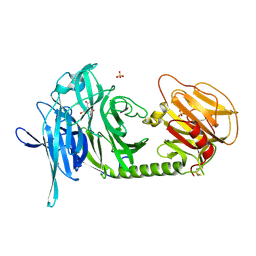 | | Trypanosoma rangeli sialidase in complex with DANA at 1.6 A | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SULFATE ION, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-20 | | Release date: | 2003-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structures of free and
inhibitor-bound Trypanosoma rangeli
sialidase and its comparison with T. cruzi
trans-sialidase
J.Mol.Biol., 325, 2003
|
|
1N1Y
 
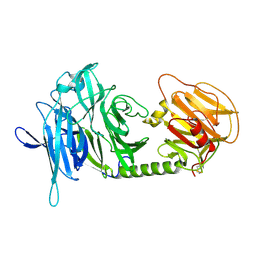 | | Trypanosoma rangeli sialidase in complex with sialic acid | | Descriptor: | N-acetyl-alpha-neuraminic acid, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-21 | | Release date: | 2003-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution structures of free and inhibitor-bound
Trypanosoma rangeli sialidase and its comparison with T.
cruzi trans-sialidase
J.Mol.Biol., 325, 2003
|
|
3TZD
 
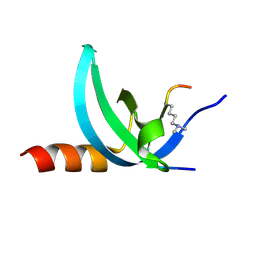 | | Crystal structure of the complex of Human Chromobox Homolog 3 (CBX3) | | Descriptor: | Chromobox protein homolog 3, Histone H1.4 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-07 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
2WHV
 
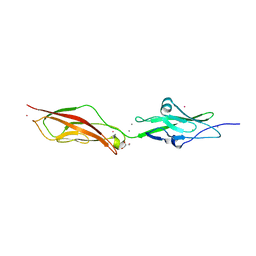 | | CRYSTAL STRUCTURE OF MOUSE CADHERIN-23 EC1-2 (ALL CATION BINDING SITES OCCUPIED BY CALCIUM) | | Descriptor: | CADHERIN-23, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Determinants of Cadherin-23 Function in Hearing and Deafness.
Neuron, 66, 2010
|
|
3C33
 
 | |
3C35
 
 | |
3C32
 
 | |
3C31
 
 | |
3C34
 
 | |
2AH2
 
 | | Trypanosoma cruzi trans-sialidase in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase
Structure, 12, 2004
|
|
3DB5
 
 | | Crystal structure of methyltransferase domain of human PR domain-containing protein 4 | | Descriptor: | PR domain zinc finger protein 4 | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of methyltransferase domain of human PR domain-containing protein 4.
To be Published
|
|
3C36
 
 | |
3DAL
 
 | | Methyltransferase domain of human PR domain-containing protein 1 | | Descriptor: | PR domain zinc finger protein 1 | | Authors: | Amaya, M.F, Zeng, H, Antoshenko, T, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-29 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure
of methyltransferase domain of human PR domain-containing protein 1
To be Published
|
|
3E0C
 
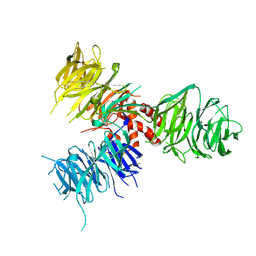 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | Descriptor: | DNA damage-binding protein 1 | | Authors: | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
2I0B
 
 | |
3F2K
 
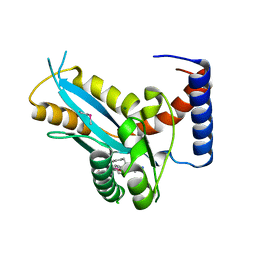 | | Structure of the transposase domain of human Histone-lysine N-methyltransferase SETMAR | | Descriptor: | Histone-lysine N-methyltransferase SETMAR, LYFA Peptide, MAGNESIUM ION | | Authors: | Amaya, M.F, Dombrovski, L, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Transposase Domain of Human
Histone-lysine N-methyltransferase SETMAR.
To be Published
|
|
3FDR
 
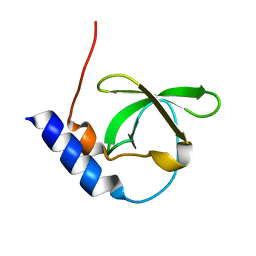 | | Crystal structure of TDRD2 | | Descriptor: | Tudor and KH domain-containing protein | | Authors: | Amaya, M.F, Adams, M.A, Guo, Y, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WD0
 
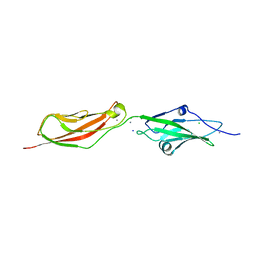 | | CRYSTAL STRUCTURE OF NONSYNDROMIC DEAFNESS (DFNB12) ASSOCIATED MUTANT D124G OF MOUSE CADHERIN-23 EC1-2 | | Descriptor: | CADHERIN-23, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2009-03-18 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Determinants of Cadherin-23 Function in Hearing and Deafness.
Neuron, 66, 2010
|
|
2F36
 
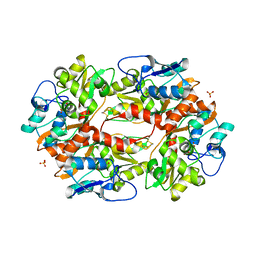 | |
3I8Z
 
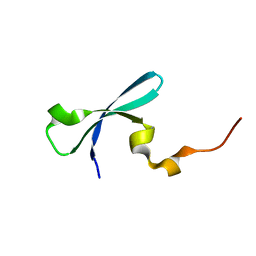 | | Crystal structure of human chromobox homolog 4 (CBX4) | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Amaya, M.F, Zhihong, L, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of human chromobox homolog 4 (CBX4)
To be Published
|
|
2K1L
 
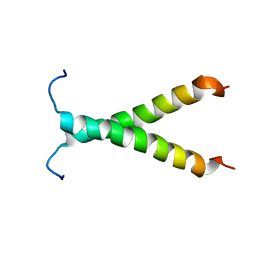 | |
