8FEJ
 
 | |
8FEL
 
 | |
7THE
 
 | | Structure of RBD directed antibody DH1042 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1042 Fab Heavy Chain, DH1042 Fab Light Chain, ... | | Authors: | May, A.J, Manne, K, Acharya, P. | | Deposit date: | 2022-01-10 | | Release date: | 2022-02-16 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural diversity of the SARS-CoV-2 Omicron spike.
Mol.Cell, 82, 2022
|
|
8DTK
 
 | |
1QFO
 
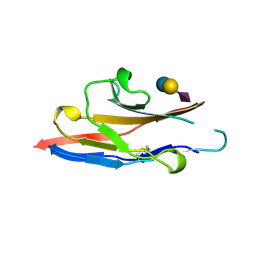 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH 3'SIALYLLACTOSE | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Vinson, M, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
1QDN
 
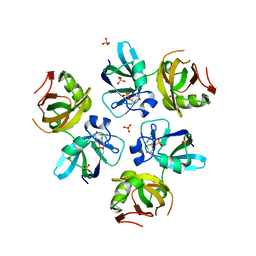 | | AMINO TERMINAL DOMAIN OF THE N-ETHYLMALEIMIDE SENSITIVE FUSION PROTEIN (NSF) | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (N-ETHYLMALEIMIDE SENSITIVE FUSION PROTEIN (NSF)), SULFATE ION | | Authors: | May, A.P, Misura, K.M.S, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1999-05-21 | | Release date: | 1999-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein.
Nat.Cell Biol., 1, 1999
|
|
1QFP
 
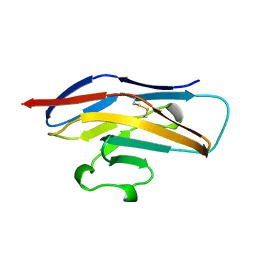 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) | | Descriptor: | PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Burtnick, L, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
8I6V
 
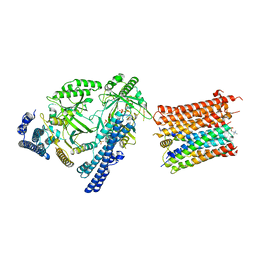 | | Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc4/Vtc3/Vtc1) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mayer, A, Wu, S, Ye, S. | | Deposit date: | 2023-01-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the polyphosphate polymerase VTC reveals coupling of polymer synthesis to membrane transit.
Embo J., 42, 2023
|
|
6S6E
 
 | |
1N37
 
 | |
7OMD
 
 | | Crystal structure of azacoelenterazine-bound Renilla reniformis luciferase variant RLuc8-D162A | | Descriptor: | 6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-8-(phenylmethyl)-[1,2,4]triazolo[4,3-a]pyrazin-3-one, CHLORIDE ION, Coelenterazine h 2-monooxygenase, ... | | Authors: | Schenkmayerova, A, Janin, Y.L, Marek, M. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
7OMR
 
 | |
7OME
 
 | |
7OMO
 
 | |
6S97
 
 | |
1S4G
 
 | | Somatomedin-B Domain of human plasma vitronectin. | | Descriptor: | HYDROXIDE ION, Vitronectin | | Authors: | Mayasundari, A, Whittemore, N.A, Serpersu, E.H, Peterson, C.B. | | Deposit date: | 2004-01-16 | | Release date: | 2004-06-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of human vitronectin: proximal sites that regulate fibrinolysis and cell migration
J.Biol.Chem., 279, 2004
|
|
7OX8
 
 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX9
 
 | | Target-bound SpCas9 complex with AAVS1 all-RNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Pacesa, M, Donohoue, P, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7THT
 
 | | CryoEM structure of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1042 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1042 heavy chain, ... | | Authors: | Manne, K, May, A, Acharya, P. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural diversity of the SARS-CoV-2 Omicron spike.
Mol.Cell, 82, 2022
|
|
8DPZ
 
 | |
3NUH
 
 | |
3NBU
 
 | | Crystal structure of pGI glucosephosphate isomerase | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase | | Authors: | Alber, T, Zubieta, C, Totir, M, May, A, Echols, N. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3N6Q
 
 | | Crystal structure of YghZ from E. coli | | Descriptor: | MAGNESIUM ION, YghZ aldo-keto reductase | | Authors: | Zubieta, C, Totir, M, Echols, N, May, A, Alber, T. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
