1QZM
 
 | | alpha-domain of ATPase | | Descriptor: | ATP-dependent protease La | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Khalatova, A.G, Rasulova, F.S, Tropea, J.E, Maurizi, M.R, Rotanova, T.V, Gustchina, A, Wlodawer, A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the AAA+ alpha domain of E. coli Lon protease at 1.9A resolution.
J.Struct.Biol., 146
|
|
2ANE
 
 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
2FZS
 
 | |
1MBV
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN HETERODIMER TETRAGONAL FORM | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, Protein yljA | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of AAA+ Chaperone ClpA
J.Biol.Chem., 277, 2002
|
|
1MBX
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN WITH TRANSITION METAL ION BOUND | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
1TG6
 
 | | Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GLYCEROL, ... | | Authors: | Kang, S.G, Maurizi, M.R, Thompson, M, Mueser, T, Ahvazi, B. | | Deposit date: | 2004-05-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP
J.Struct.Biol., 148, 2004
|
|
1R6B
 
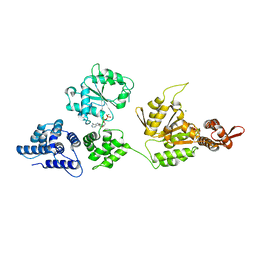 | | High resolution crystal structure of ClpA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ClpA protein, MAGNESIUM ION | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
1KSF
 
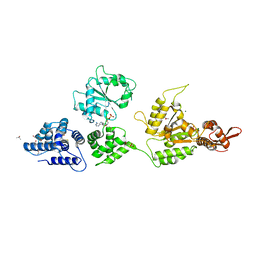 | | Crystal Structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease: Structural basis of differences in Function of the Two AAA+ ATPase domains | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT CLP PROTEASE ATP-BINDING SUBUNIT CLPA, ISOPROPYL ALCOHOL, ... | | Authors: | Guo, F, Maurizi, M.R, Esser, L, Xia, D. | | Deposit date: | 2002-01-12 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease
J.Biol.Chem., 277, 2002
|
|
1K6K
 
 | | Crystal Structure of ClpA, an AAA+ Chaperone-like Regulator of ClpAP protease implication to the functional difference of two ATPase domains | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ATP-BINDING SUBUNIT CLPA | | Authors: | Guo, F, Maurizi, M.R, Esser, L, Xia, D. | | Deposit date: | 2001-10-16 | | Release date: | 2002-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease
J.Biol.Chem., 277, 2002
|
|
1MBU
 
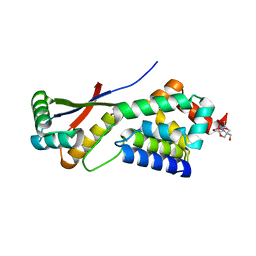 | | Crystal Structure Analysis of ClpSN heterodimer | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
1R6C
 
 | | High resolution structure of ClpN | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
1RRE
 
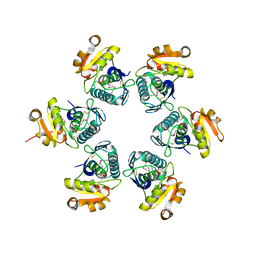 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1R6Q
 
 | | ClpNS with fragments | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-16 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1R6O
 
 | | ATP-dependent Clp protease ATP-binding subunit clpA/ATP-dependent Clp protease adaptor protein clpS | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
