3ORC
 
 | |
1THL
 
 | | Thermolysin complexed with a novel glutaramide derivative, n-(1-(2(r,s)-carboxy-4-phenylbutyl) cyclopentylcarbonyl)-(s)-tryptophan | | 分子名称: | CALCIUM ION, N-({1-[(2S)-2-carboxy-4-phenylbutyl]cyclopentyl}carbonyl)-L-tryptophan, THERMOLYSIN, ... | | 著者 | Holland, D.R, Matthews, B.W. | | 登録日 | 1993-11-17 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Inhibition of Thermolysin and Neutral Endopeptidase 24.11 By a Novel Glutaramide Derivative; X-Ray Structure Determination of the Thermolysin-Inhibitor Complex
Biochemistry, 33, 1994
|
|
4BCL
 
 | |
3MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE TRANSITION-STATE INHIBITOR COMPLEX | | 分子名称: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, SODIUM ION, ... | | 著者 | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | 登録日 | 1999-03-29 | | 公開日 | 1999-06-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
2TMN
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | 分子名称: | CALCIUM ION, N~2~-phosphono-L-leucinamide, Thermolysin, ... | | 著者 | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | 登録日 | 1987-06-29 | | 公開日 | 1989-01-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
4MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE HIS79ALA MUTANT | | 分子名称: | PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | 著者 | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | 登録日 | 1999-03-29 | | 公開日 | 1999-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
200L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | 登録日 | 1995-11-06 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
258L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, ZINC ION | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-01-05 | | 公開日 | 2000-09-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
260L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, NICKEL (II) ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-03-01 | | 公開日 | 2000-09-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
259L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, COBALT (II) ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-02-10 | | 公開日 | 1999-04-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
257L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-01-05 | | 公開日 | 2000-09-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
1P5C
 
 | |
1P56
 
 | |
221L
 
 | |
224L
 
 | |
1QUG
 
 | | E108V MUTANT OF T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | 登録日 | 1999-07-01 | | 公開日 | 1999-07-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
1QSQ
 
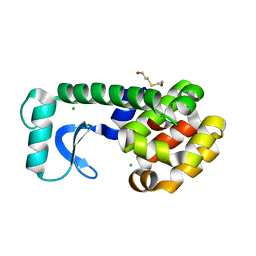 | | CAVITY CREATING MUTATION | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Lindstrom, J, Matthews, B.W. | | 登録日 | 1999-06-22 | | 公開日 | 1999-06-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1QT5
 
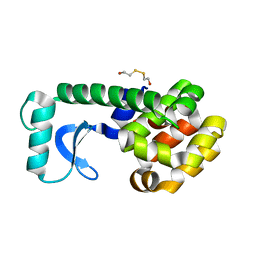 | |
2LZM
 
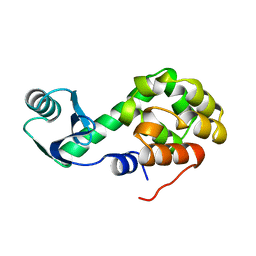 | |
1QT4
 
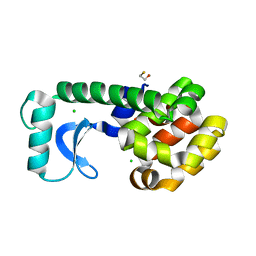 | | T26Q MUTANT OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | 著者 | Kuroki, R, Weaver, L.H, Matthews, B.W. | | 登録日 | 1999-06-30 | | 公開日 | 1999-07-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT6
 
 | | E11H Mutant of T4 Lysozyme | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | 著者 | Kuroki, R, Weaver, L.H, Matthews, B.W. | | 登録日 | 1999-06-30 | | 公開日 | 1999-07-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1T8A
 
 | | USE OF SEQUENCE DUPLICATION TO ENGINEER A LIGAND-TRIGGERED LONG-DISTANCE MOLECULAR SWITCH IN T4 Lysozyme | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, GUANIDINE, ... | | 著者 | Yousef, M.S, Baase, W.A, Matthews, B.W. | | 登録日 | 2004-05-11 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T97
 
 | |
1MAT
 
 | |
1CVK
 
 | | T4 LYSOZYME MUTANT L118A | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | 登録日 | 1999-08-23 | | 公開日 | 1999-11-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
