4V4O
 
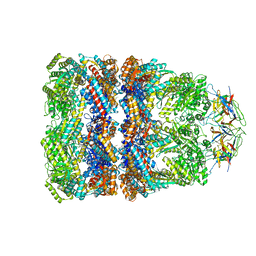 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
1D2M
 
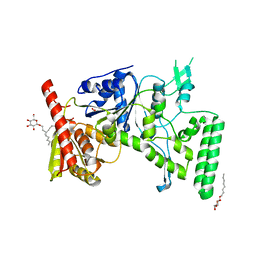 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
2GGS
 
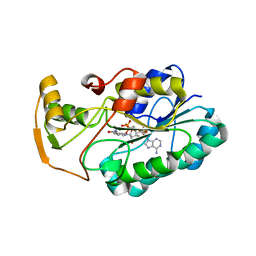 | | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii | | Descriptor: | 273aa long hypothetical dTDP-4-dehydrorhamnose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii
To be published
|
|
7CKJ
 
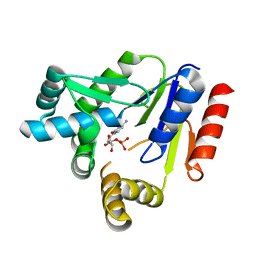 | | Crystal structure of CMP kinase in complex with CMP from Thermus thermophilus HB8 | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Cytidylate kinase | | Authors: | Mega, R, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of Thermus thermophilus CMP kinase complexed with a phosphoryl group acceptor and donor.
PLoS ONE, 15, 2020
|
|
5H45
 
 | | Crystal structure of the C-terminal Lon protease-like domain of Thermus thermophilus RadA/Sms | | Descriptor: | DNA repair protein RadA | | Authors: | Inoue, M, Fukui, K, Fujii, Y, Nakagawa, N, Yano, T, Kuramitsu, S, Masui, R. | | Deposit date: | 2016-10-30 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lon protease-like domain in the bacterial RecA paralog RadA is required for DNA binding and repair.
J. Biol. Chem., 292, 2017
|
|
1ONL
 
 | | Crystal structure of Thermus thermophilus HB8 H-protein of the glycine cleavage system | | Descriptor: | glycine cleavage system H protein | | Authors: | Nakai, T, Ishijima, J, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-28 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Thermus thermophilus HB8 H-protein of the glycine-cleavage system, resolved by a six-dimensional molecular-replacement method.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3HJE
 
 | | Crystal structure of sulfolobus tokodaii hypothetical maltooligosyl trehalose synthase | | Descriptor: | 704aa long hypothetical glycosyltransferase, GLYCEROL | | Authors: | Cielo, C.B.C, Okazaki, S, Suzuki, A, Mizushima, T, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2009-05-21 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ST0929, a putative glycosyl transferase from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2YVO
 
 | | Crystal structure of NDX2 in complex with MG2+ and AMP from thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MutT/nudix family protein | | Authors: | Wakamatsu, T, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8
J.Bacteriol., 190, 2008
|
|
2YYB
 
 | | Crystal structure of TTHA1606 from Thermus thermophilus HB8 | | Descriptor: | Hypothetical protein TTHA1606 | | Authors: | Tomoike, F, Nakagwa, N, Ebihara, A, Yokoyama, S, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-28 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the conserved hypothetical protein TTHA1606 from Thermus thermophilus HB8.
Proteins, 76, 2009
|
|
7ED9
 
 | | Crystal structure of selenomethionine-labeled Thermus thermophilus FakA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable kinase | | Authors: | Nakatani, M, Nakahara, S, Fukui, K, Murakawa, T, Masui, R. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01764154 Å) | | Cite: | Crystal structure of a nucleotide-binding domain of fatty acid kinase FakA from Thermus thermophilus HB8.
J.Struct.Biol., 214, 2022
|
|
7ED6
 
 | | Crystal structure of Thermus thermophilus FakA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable kinase | | Authors: | Nakatani, M, Nakahara, S, Fukui, K, Murakawa, T, Masui, R. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-01-11 | | Method: | X-RAY DIFFRACTION (1.92850327 Å) | | Cite: | Crystal structure of a nucleotide-binding domain of fatty acid kinase FakA from Thermus thermophilus HB8.
J.Struct.Biol., 214, 2022
|
|
1IR6
 
 | | Crystal structure of exonuclease RecJ bound to manganese | | Descriptor: | MANGANESE (II) ION, exonuclease RecJ | | Authors: | Yamagata, A, Kakuta, Y, Masui, R, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5YTQ
 
 | | Crystal Structure of TTHA0139 L34A with Lanthanum from Thermus thermophilus HB8 | | Descriptor: | LANTHANUM (III) ION, TTHA0139 | | Authors: | Takao, K, Inoue, M, Fukui, K, Yano, T, Masui, R. | | Deposit date: | 2017-11-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal Structure of TTHA0139 L34A with Lanthanum from Thermus thermophilus HB8
To Be Published
|
|
5YTP
 
 | |
1IQU
 
 | | Crystal structure of photolyase-thymine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, THYMINE, ... | | Authors: | Komori, H, Masui, R, Kuramitsu, S, Yokoyama, S, Shibata, T, Inoue, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-08-03 | | Release date: | 2002-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IQR
 
 | | Crystal structure of DNA photolyase from Thermus thermophilus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, photolyase | | Authors: | Komori, H, Masui, R, Kuramitsu, S, Yokoyama, S, Shibata, T, Inoue, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-07-27 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3W8N
 
 | |
3W90
 
 | |
3IEK
 
 | | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IEL
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP
To be Published
|
|
3IE1
 
 | | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA | | Descriptor: | CITRATE ANION, RNA (5'-R(P*UP*UP*UP*U)-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA
To be Published
|
|
3IE2
 
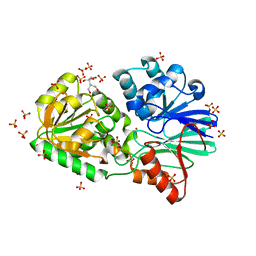 | | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IDZ
 
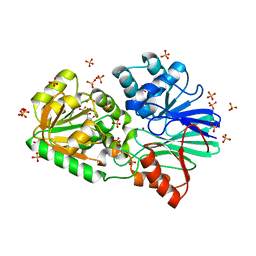 | | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IE0
 
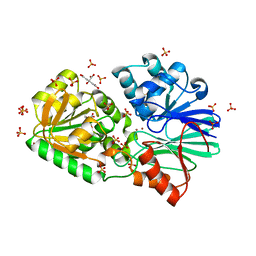 | | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IEM
 
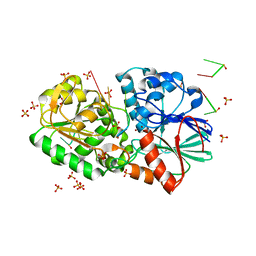 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog | | Descriptor: | CITRATE ANION, RNA (5'-R(*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU))-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitus, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog
To be Published
|
|
