7TT9
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y34F variant | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, J.E, Liu, K, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y34F variant
To Be Published
|
|
1BK2
 
 | |
6CII
 
 | |
5A66
 
 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and manganese ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A67
 
 | |
5A68
 
 | |
5A65
 
 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine diphosphate, orthophosphate and magnesium ions. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A64
 
 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine triphosphate. | | Descriptor: | 1,2-ETHANEDIOL, THIAMINE TRIPHOSPHATASE, TRIETHYLENE GLYCOL, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A60
 
 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two magnesium ions | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A61
 
 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two manganese ions. | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A5Y
 
 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
2YI1
 
 | | Crystal structure of N-Acetylmannosamine kinase in complex with N- acetyl mannosamine 6-phosphate and ADP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
2YHW
 
 | | High-resolution crystal structures of N-Acetylmannosamine kinase: Insights about substrate specificity, activity and inhibitor modelling. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ACETATE ION, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Zimmer, R, Tauberger, E, Reutter, W, Saenger, W, Fan, H, Moniot, S. | | Deposit date: | 2011-05-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
2YHY
 
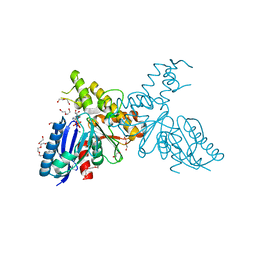 | | Structure of N-Acetylmannosamine kinase in complex with N- acetylmannosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
1PWT
 
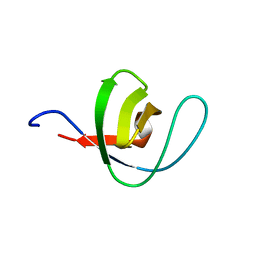 | | THERMODYNAMIC ANALYSIS OF ALPHA-SPECTRIN SH3 AND TWO OF ITS CIRCULAR PERMUTANTS WITH DIFFERENT LOOP LENGTHS: DISCERNING THE REASONS FOR RAPID FOLDING IN PROTEINS | | Descriptor: | ALPHA SPECTRIN | | Authors: | Martinez, J.C, Viguera, A.R, Berisio, R, Wilmanns, M, Mateo, P.L, Filmonov, V.V, Serrano, L. | | Deposit date: | 1998-10-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermodynamic analysis of alpha-spectrin SH3 and two of its circular permutants with different loop lengths: discerning the reasons for rapid folding in proteins.
Biochemistry, 38, 1999
|
|
6ZK7
 
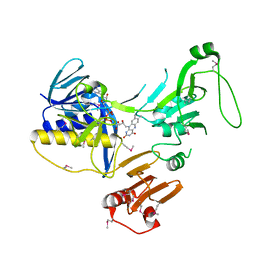 | | Crystal Structure of human PYROXD1/FAD complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 1 | | Authors: | Meinhart, A, Asanovic, I, Martinez, J, Clausen, T. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The oxidoreductase PYROXD1 uses NAD(P) + as an antioxidant to sustain tRNA ligase activity in pre-tRNA splicing and unfolded protein response.
Mol.Cell, 81, 2021
|
|
5C9B
 
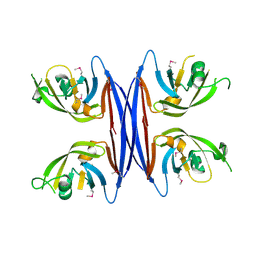 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5C9D
 
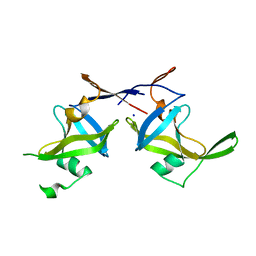 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8UGZ
 
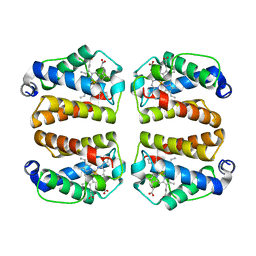 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heme d formation in a Shewanella benthica hemoglobin.
J.Inorg.Biochem., 259, 2024
|
|
8VSH
 
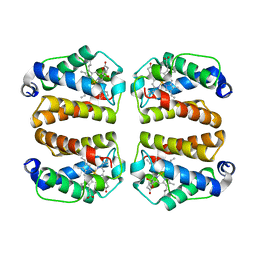 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant with trans heme D | | Descriptor: | Group 1 truncated hemoglobin, {3-[(2R,5'R)-9',14'-diethenyl-5'-hydroxy-5',10',15',19'-tetramethyl-5-oxo-4,5-dihydro-3H-spiro[furan-2,4'-[21,22,23,24]tetraazapentacyclo[16.2.1.13,6.18,11.113,16]tetracosa[1,3(24),6,8,10,12,14,16(22),17,19]decaen]-20'-yl-kappa~4~N~21'~,N~22'~,N~23'~,N~24'~]propanoato}iron | | Authors: | Lecomte, J.T.J, Martinez, J.E, Schlessman, J.L, Schultz, T.D, Siegler, M.A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Heme d formation in a Shewanella benthica hemoglobin.
J.Inorg.Biochem., 259, 2024
|
|
8UZU
 
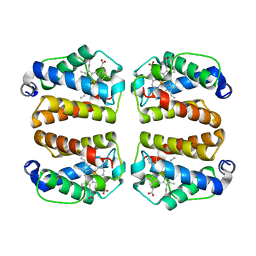 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin L80A C51S C71S variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin L80A C51S C71S variant
To Be Published
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
7Z67
 
 | |
