1OHN
 
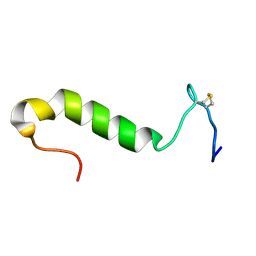 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-05-28 | | Release date: | 2003-09-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
2BOW
 
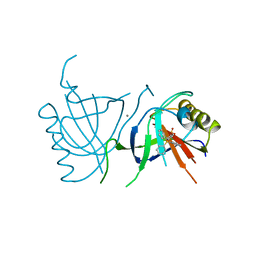 | | MULTIDRUG-BINDING DOMAIN OF TRANSCRIPTION ACTIVATOR BMRR IN COMPLEX WITH A LIGAND, TETRAPHENYLPHOSPHONIUM | | Descriptor: | MANGANESE (II) ION, MULTIDRUG-EFFLUX TRANSPORTER 1 REGULATOR BMRR, SULFATE ION, ... | | Authors: | Zheleznova, E.E, Markham, P.N, Neyfakh, A.A, Brennan, R.G. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter.
Cell(Cambridge,Mass.), 96, 1999
|
|
1Q5K
 
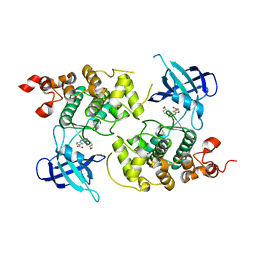 | | crystal structure of Glycogen synthase kinase 3 in complexed with inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-(4-METHOXYBENZYL)-N'-(5-NITRO-1,3-THIAZOL-2-YL)UREA | | Authors: | Bhat, R, Xue, Y, Berg, S, Hellberg, S, Ormo, M, Nilsson, Y, Radesater, A.C, Jerning, E, Markgren, P.O, Borgegard, T, Nylof, M, Gimenez-Cassina, A, Hernandez, F, Lucas, J.J, Diaz-Mido, J, Avila, J. | | Deposit date: | 2003-08-08 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418.
J.Biol.Chem., 278, 2003
|
|
1SYY
 
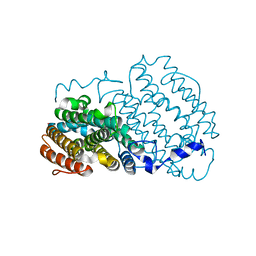 | | Crystal structure of the R2 subunit of ribonucleotide reductase from Chlamydia trachomatis | | Descriptor: | FE (III) ION, LEAD (II) ION, Ribonucleoside-diphosphate reductase beta chain | | Authors: | Hogbom, M, Stenmark, P, Voevodskaya, N, McClarty, G, Graslund, A, Nordlund, P. | | Deposit date: | 2004-04-02 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The radical site in chlamydial ribonucleotide reductase defines a new R2 subclass.
Science, 305, 2004
|
|
2OBC
 
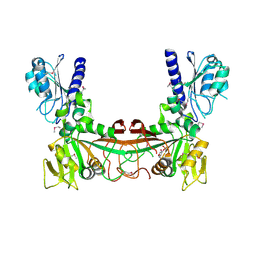 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2O7P
 
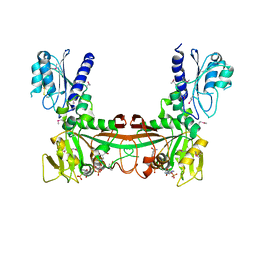 | | The crystal structure of RibD from Escherichia coli in complex with the oxidised NADP+ cofactor in the active site of the reductase domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2L49
 
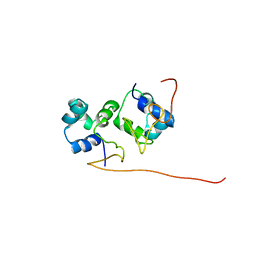 | | The solution structure of the P2 C,the immunity repressor of the P2 bacteriophage | | Descriptor: | C protein | | Authors: | Massad, T, Papadopolos, E, Stenmark, P, Damberg, P. | | Deposit date: | 2010-10-01 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The C repressor of the P2 bacteriophage.
J.Biomol.Nmr, 64, 2016
|
|
2NMQ
 
 | |
2HW4
 
 | | Crystal structure of human phosphohistidine phosphatase | | Descriptor: | 14 kDa phosphohistidine phosphatase, FORMIC ACID | | Authors: | Busam, R.D, Thorsell, A.G, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-31 | | Release date: | 2006-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First structure of a eukaryotic phosphohistidine phosphatase
J.Biol.Chem., 281, 2006
|
|
2MFZ
 
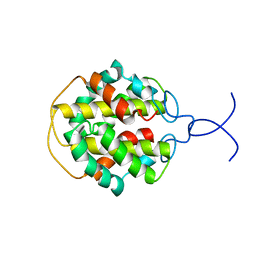 | | NMR structure of C-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Andersson, M, Chen, G, Landreh, M, Nordling, K, Kronqvist, N, Westermark, P, Jornvall, H, Knight, S, Ridderstrale, Y, Holm, L, Meng, Q, Chesler, M, Johansson, J, Rising, A. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains
Plos Biol., 12, 2014
|
|
