3HDS
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methylmuconolactone methylisomerase, short peptide ASWSA | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-07 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
3HFK
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase (H52A) in complex with 4-methylmuconolactone | | Descriptor: | 4-methylmuconolactone methylisomerase, [(2S)-2-methyl-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
3HF5
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase in complex with 3-methylmuconolactone | | Descriptor: | 4-methylmuconolactone methylisomerase, [(2S)-3-methyl-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
2GVQ
 
 | | Anthranilate phosphoribosyl-transferase (TRPD) from S. solfataricus in complex with anthranilate | | Descriptor: | 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2006-05-03 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
1M9S
 
 | | Crystal structure of Internalin B (InlB), a Listeria monocytogenes virulence protein containing SH3-like domains. | | Descriptor: | Internalin B, SULFATE ION, TERBIUM(III) ION | | Authors: | Marino, M, Banerjee, M, Jonquieres, R, Cossart, P, Ghosh, P. | | Deposit date: | 2002-07-29 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands
Embo J., 21, 2002
|
|
1ZYK
 
 | | Anthranilate Phosphoribosyltransferase in complex with PRPP, anthranilate and magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase, ... | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
1ZXY
 
 | | Anthranilate Phosphoribosyltransferase from Sulfolobus solfataricus in complex with PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2005-06-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
2A38
 
 | | Crystal structure of the N-Terminus of titin | | Descriptor: | CADMIUM ION, Titin | | Authors: | Marino, M, Muhle-Goll, C, Svergun, D, Demirel, M, Mayans, O. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ig doublet Z1Z2: a model system for the hybrid analysis of conformational dynamics in Ig tandems from titin
Structure, 14, 2006
|
|
1OTO
 
 | | Calcium-binding mutant of the internalin B LRR domain | | Descriptor: | CALCIUM ION, Internalin B | | Authors: | Marino, M, Copp, J, Dramsi, S, Chapman, T, van der Geer, P, Cossart, P, Ghosh, P. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterization of the calcium-binding sites of Listeria monocytogenes InlB
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1OTN
 
 | | Calcium-binding mutant of the Internalin B LRR domain | | Descriptor: | CALCIUM ION, Internalin B | | Authors: | Marino, M, Copp, J, Dramsi, S, Chapman, T, van der Geer, P, Cossart, P, Ghosh, P. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the calcium-binding sites of Listeria monocytogenes InlB
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1OTM
 
 | | Calcium-binding mutant of the internalin B LRR domain | | Descriptor: | internalin B | | Authors: | Marino, M, Copp, J, Dramsi, S, Chapman, T, van der Geer, P, Cossart, P, Ghosh, P. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of the calcium-binding sites of Listeria monocytogenes InlB
Biochem.Biophys.Res.Commun., 316, 2004
|
|
2RIK
 
 | | I-band fragment I67-I69 from titin | | Descriptor: | Titin | | Authors: | Marino, M, von Castelmur, E, Labeit, D, Labeit, S, Mayans, O. | | Deposit date: | 2007-10-11 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1D0B
 
 | | INTERNALIN B LEUCINE RICH REPEAT DOMAIN | | Descriptor: | CALCIUM ION, INTERNALIN B | | Authors: | Marino, M, Braun, L, Cossart, P, Ghosh, P. | | Deposit date: | 1999-09-09 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the lnlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes.
Mol.Cell, 4, 1999
|
|
2JML
 
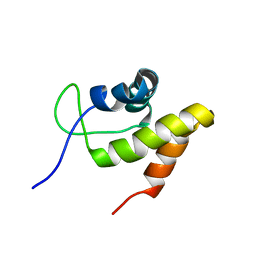 | | Solution structure of the N-terminal domain of CarA repressor | | Descriptor: | DNA BINDING DOMAIN/TRANSCRIPTIONAL REGULATOR | | Authors: | Jimenez, M, Padmanabhan, S, Gonzalez, C, Perez-Marin, M.C, Elias-Arnanz, M, Murillo, F.J, Rico, M. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for operator and antirepressor recognition by Myxococcus xanthus CarA repressor.
Mol.Microbiol., 63, 2007
|
|
8XI6
 
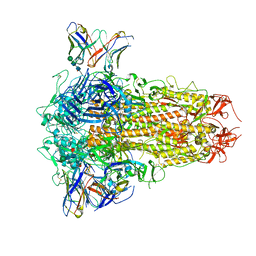 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 98, 2024
|
|
8BBS
 
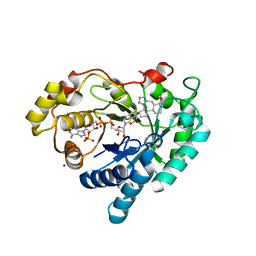 | | Structure of AKR1C3 in complex with a bile acid fused tetrazole inhibitor | | Descriptor: | (4~{R})-4-[(1~{R},2~{S},5~{R},6~{R},13~{S},14~{S},17~{R},19~{R})-6,14-dimethyl-17-oxidanyl-7,8,9,10-tetrazapentacyclo[11.8.0.0^{2,6}.0^{7,11}.0^{14,19}]henicosa-8,10-dien-5-yl]pentanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Petri, E.T, Skerlova, J, Marinovic, M, Brynda, J, Kugler, M, Skoric, D, Bekic, S, Celic, A.S, Rezacova, P. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of human aldo-keto reductase 1C3 in complex with a bile acid fused tetrazole inhibitor: experimental validation, molecular docking and structural analysis.
Rsc Med Chem, 14, 2023
|
|
3B43
 
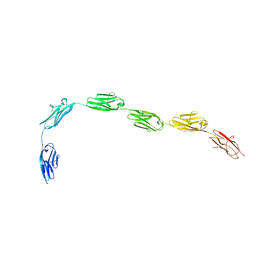 | | I-band fragment I65-I70 from titin | | Descriptor: | Titin | | Authors: | von Castelmur, E, Marino, M, Labeit, D, Labeit, S, Mayans, O. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ZLJ
 
 | | CRYSTAL STRUCTURE OF FULL-LENGTH E.COLI DNA MISMATCH REPAIR PROTEIN MUTS D835R MUTANT IN COMPLEX WITH GT MISMATCHED DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP *AP*GP*TP*GP*TP*CP*AP)-3', 5'-D(*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*TP)-3', DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Groothuizen, F.S, Fish, A, Petoukhov, M.V, Reumer, A, Manelyte, L, Winterwerp, H.H.K, Marinus, M.G, Lebbink, J.H.G, Svergun, D.I, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2013-02-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation.
Nucleic Acids Res., 41, 2013
|
|
1WT6
 
 | | Coiled-Coil domain of DMPK | | Descriptor: | Myotonin-protein kinase | | Authors: | Garcia, P, Marino, M, Mayans, O. | | Deposit date: | 2004-11-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into the self-assembly mechanism of dystrophia myotonica kinase
Faseb J., 20, 2006
|
|
2RJM
 
 | | 3Ig structure of titin domains I67-I69 E-to-A mutated variant | | Descriptor: | Titin | | Authors: | von Castelmur, E, Marino, M, Labeit, D, Labeit, S, Mayans, O. | | Deposit date: | 2007-10-15 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2V4X
 
 | | Crystal Structure of Jaagsiekte Sheep Retrovirus Capsid N-terminal domain | | Descriptor: | CAPSID PROTEIN P27 | | Authors: | Mortuza, G.B, Goldstone, D.C, Pashley, C, Haire, L.F, Palmarini, M, Taylor, W.R, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Capsid Amino-Terminal Domain from the Betaretrovirus, Jaagsiekte Sheep Retrovirus.
J.Mol.Biol., 386, 2009
|
|
