3ZNV
 
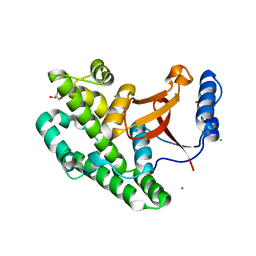 | | Crystal structure of the OTU domain of OTULIN at 1.3 Angstroms. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
6NKK
 
 | | Structure of PhqE Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and premalbrancheamide | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
1P83
 
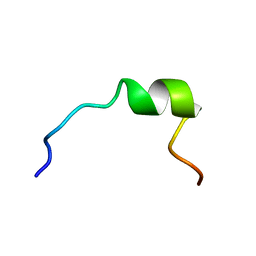 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
4A7F
 
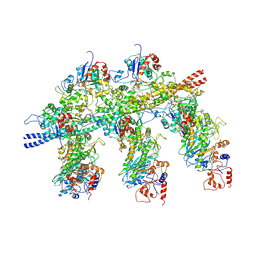 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 3) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
1T5G
 
 | | Arginase-F2-L-Arginine complex | | Descriptor: | ARGININE, Arginase 1, FLUORIDE ION, ... | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Han, S, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-05-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
1TH9
 
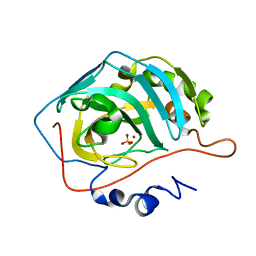 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, SULFATE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-06-01 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1SZX
 
 | | Role Of Hydrogen Bonding In The Active Site Of Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Greenleaf, W.B, Perry, J.J, Hearn, A.S, Cabelli, D.E, Lepock, J.R, Stroupe, M.E, Tainer, J.A, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-04-06 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of hydrogen bonding in the active site of human manganese superoxide dismutase.
Biochemistry, 43, 2004
|
|
1NYH
 
 | | Crystal Structure of the Coiled-coil Dimerization Motif of Sir4 | | Descriptor: | Regulatory protein SIR4 | | Authors: | Chang, J.F, Hall, B.E, Tanny, J.C, Moazed, D, Filman, D, Ellenberger, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Coiled-coil Dimerization Motif of Sir4 and Its Interaction With Sir3
Structure, 11, 2003
|
|
3SS1
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain | | Descriptor: | MANGANESE (II) ION, Toxin A | | Authors: | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2011-07-07 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
2AYR
 
 | | A SERM Designed for the Treatment of Uterine Leiomyoma with Unique Tissue Specificity for Uterus and Ovaries in Rats | | Descriptor: | 6-(4-METHYLSULFONYL-PHENYL)-5-[4-(2-PIPERIDIN-1-YLETHOXY)PHENOXY]NAPHTHALEN-2-OL, Estrogen receptor | | Authors: | Hummel, C.W, Geiser, A.G, Bryant, H.U, Cohen, I.R, Dally, R.D, Fong, K.C, Frank, S.A, Hinklin, R, Jones, S.A, Lewis, G, McCann, D.J, Shepherd, T.A, Tian, H, Rudman, D.G, Wallace, O.B, Wang, Y, Dodge, J.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A selective estrogen receptor modulator designed for the treatment of uterine leiomyoma with unique tissue specificity for uterus and ovaries in rats
J.Med.Chem., 48, 2005
|
|
6NKM
 
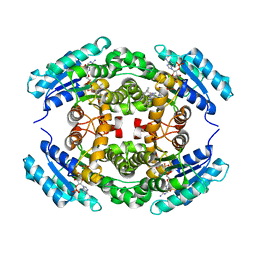 | | Structure of PhqE D166N Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and substrate | | Descriptor: | 3-{[2-(2-methylbut-3-en-2-yl)-1H-indol-3-yl]methyl}-8H-pyrrolo[1,2-a]pyrazin-5-ium-1-olate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
4A7N
 
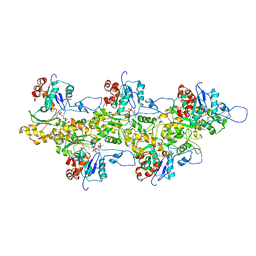 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
6NKI
 
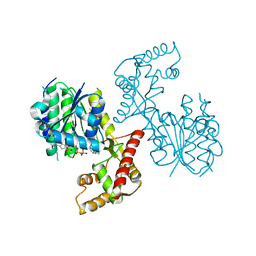 | | Structure of PhqB Reductase Domain from Penicillium fellutanum | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NRPS | | Authors: | Dan, Q, Newmister, S.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
3Q0B
 
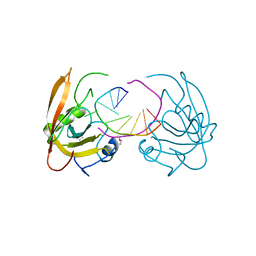 | |
3Q0F
 
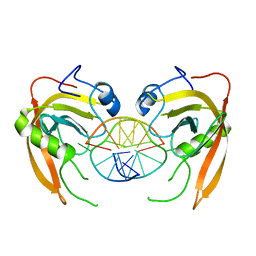 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3Q0C
 
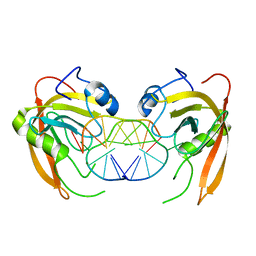 | | Crystal structure of SUVH5 SRA-fully methylated CG DNA complex in space group P6122 | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*(5CM)P*GP*TP*AP*GP*TP*T)-3'), Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6567 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3PHW
 
 | | OTU Domain of Crimean Congo Hemorrhagic Fever Virus in complex with Ubiquitin | | Descriptor: | ETHANAMINE, RNA-directed RNA polymerase L, Ubiquitin-40S ribosomal protein S27a | | Authors: | Akutsu, M, Ye, Y, Virdee, S, Komander, D. | | Deposit date: | 2010-11-04 | | Release date: | 2011-02-02 | | Last modified: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4A7H
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 2) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4LRF
 
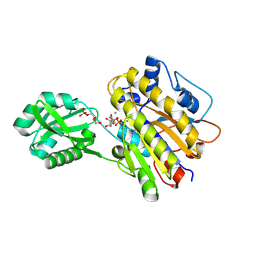 | | Phosphopentomutase S154G variant soaked with ribose 5-phosphate | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
1TME
 
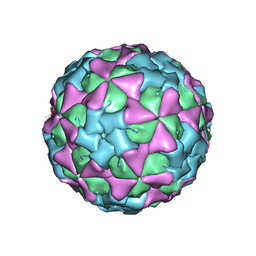 | | THREE-DIMENSIONAL STRUCTURE OF THEILER VIRUS | | Descriptor: | THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP1), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP2), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP3), ... | | Authors: | Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1992-01-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of Theiler virus.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
3SRZ
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain bound to UDP-glucose | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2011-07-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
1UXK
 
 | | Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface | | Descriptor: | CADMIUM ION, CHLORIDE ION, MALATE DEHYDROGENASE, ... | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large Improvement in the Thermal Stability of a Tetrameric Malate Dehydrogenase by Single Point Mutations at the Dimer-Dimer Interface.
J.Mol.Biol., 341, 2004
|
|
1UX4
 
 | | Crystal structures of a Formin Homology-2 domain reveal a tethered-dimer architecture | | Descriptor: | BNI1 PROTEIN | | Authors: | Xu, Y, Moseley, J.B, Sagot, I, Poy, F, Pellman, D, Goode, B.L, Eck, M.J. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture
Cell(Cambridge,Mass.), 116, 2004
|
|
1UPQ
 
 | | Crystal structure of the pleckstrin homology (PH) domain of PEPP1 | | Descriptor: | PEPP1, SULFATE ION | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
