4Q32
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(naphthalen-2-yl)-2-[2-(pyridin-2-yl)-1H-benzimidazol-1-yl]acetamide | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91
To be Published
|
|
5UWX
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P176 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P176
To Be Published
|
|
5UXE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P178 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P178
To Be Published
|
|
3UHP
 
 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | Glutamate racemase | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published, 2011
|
|
3UHF
 
 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | CHLORIDE ION, D-GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published
|
|
4R7T
 
 | | Crystal structure of glucosamine-6-phosphate deaminase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucosamine-6-phosphate deaminase from Vibrio cholerae
To be Published
|
|
4GUD
 
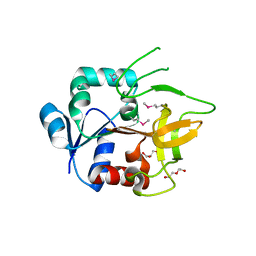 | | Crystal Structure of Amidotransferase HisH from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Crystal Structure of Amidotransferase HisH from Vibrio cholerae.
To be Published
|
|
4PWT
 
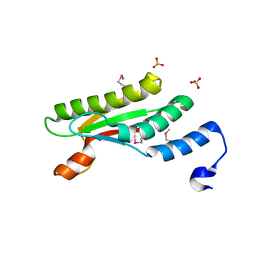 | | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from Yersinia pestis CO92 | | Descriptor: | FORMIC ACID, PYROPHOSPHATE 2-, Peptidoglycan-associated lipoprotein, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from
Yersinia pestis CO92
To be Published
|
|
4PZK
 
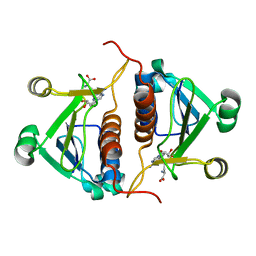 | | Crystal strucrure of putative RNA methyltransferase from Bacillus anthracis. | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (cytidine(34)-2'-O)-methyltransferase | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal strucrure of putative RNA methyltransferase from Bacillus anthracis.
To be Published
|
|
4QI9
 
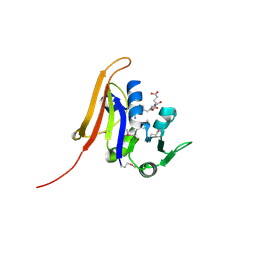 | | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate
To be Published
|
|
3Q1H
 
 | | Crystal Structure of Dihydrofolate Reductase from Yersinia pestis | | Descriptor: | Dihydrofolate reductase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Yersinia pestis
To be Published
|
|
3OS4
 
 | | The Crystal Structure of Nicotinate Phosphoribosyltransferase from Yersinia pestis | | Descriptor: | ACETIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-08 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The Crystal Structure of Nicotinate Phosphoribosyltransferase from Yersinia pestis
TO BE PUBLISHED
|
|
3OUU
 
 | | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni | | Descriptor: | Biotin carboxylase, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
3QTY
 
 | | Crystal structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis complexed with pyrophosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis complexed with pyrophosphate
To be Published
|
|
3QTT
 
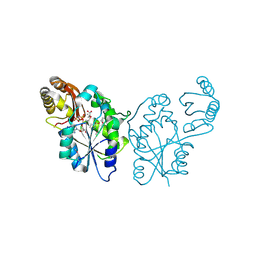 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine | | Descriptor: | BETA-ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine.
To be Published
|
|
3O6V
 
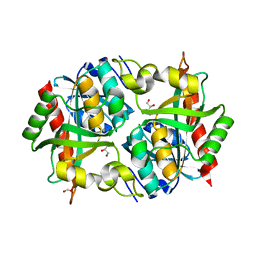 | | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | FORMIC ACID, GLYCEROL, Uridine phosphorylase | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3PEI
 
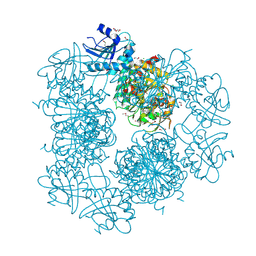 | | Crystal Structure of Cytosol Aminopeptidase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | Authors: | Maltseva, N, Kim, Y, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Cytosol Aminopeptidase from
Francisella tularensis
To be Published
|
|
3R8X
 
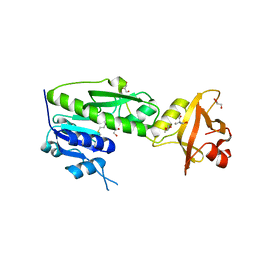 | | Crystal Structure of Methionyl-tRNA Formyltransferase from Yersinia pestis complexed with L-methionine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, METHIONINE, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Crystal Structure of Methionyl-tRNA Formyltransferase
from Yersinia pestis complexed with L-methionine
To be Published
|
|
3PNS
 
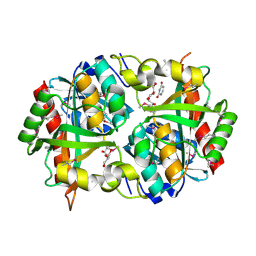 | | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3OUZ
 
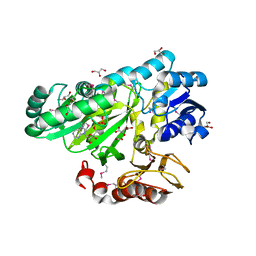 | | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, D-MALATE, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
1I6N
 
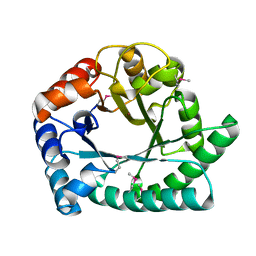 | | 1.8 A Crystal structure of IOLI protein with a binding zinc atom | | Descriptor: | IOLI PROTEIN, ZINC ION | | Authors: | Zhang, R.G, Dementiva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Alkire, R, Maltsev, N, Korolev, O, Dieckman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-02 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
5UQH
 
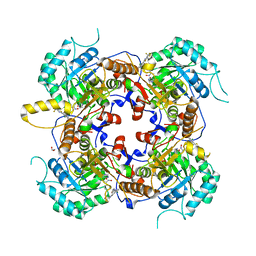 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p182 | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
5UPV
 
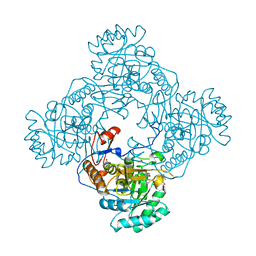 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36
To Be Published
|
|
5URQ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-alpha-D-ribofuranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176
To Be Published
|
|
5UQF
 
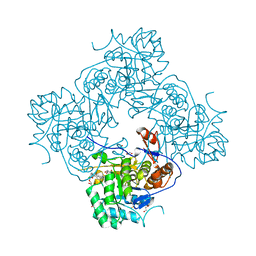 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with IMP and the inhibitor P225 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Campylobacter jejuni in the complex with IMP and the inhibitor P225
To Be Published
|
|
