4W5U
 
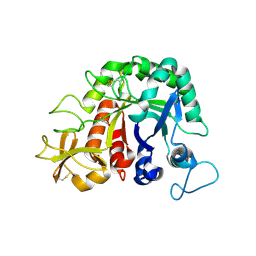 | |
4W5Z
 
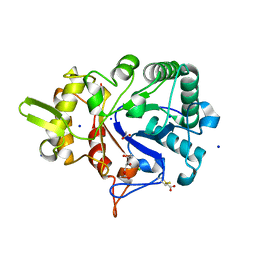 | |
6SMK
 
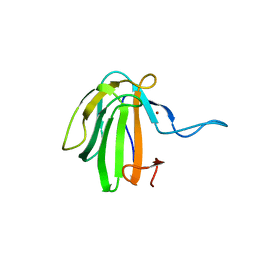 | | Crystal structure of catalytic domain A109H mutant of prophage-encoded M23 protein EnpA from Enterococcus faecalis. | | Descriptor: | Peptidase_M23 domain-containing protein, ZINC ION | | Authors: | Malecki, P.H, Mitkowski, P, Czapinska, H, Sabala, I. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Characterization of EnpA D,L-Endopeptidase from Enterococcus faecalis Prophage Provides Insights into Substrate Specificity of M23 Peptidases.
Int J Mol Sci, 22, 2021
|
|
3O5W
 
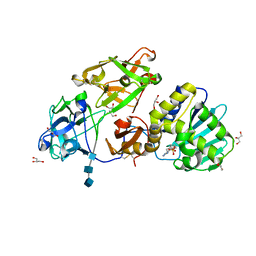 | | Binding of kinetin in the active site of mistletoe lectin I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Malecki, P.H, Meyer, A, Rypniewski, W, Szymanski, M, Barciszewski, J, Betzel, C. | | Deposit date: | 2010-07-28 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of the plant hormone kinetin in the active site of Mistletoe Lectin I from Viscum album.
Biochim.Biophys.Acta, 1824, 2012
|
|
7O5M
 
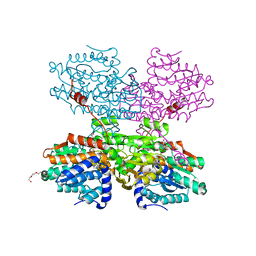 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Na+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7O5L
 
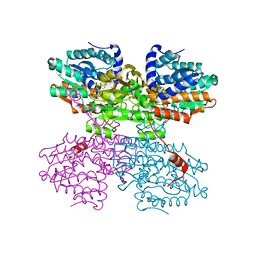 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4MB4
 
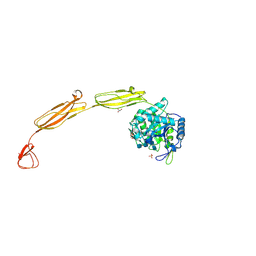 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB5
 
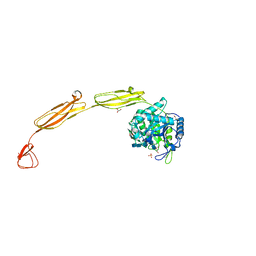 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB3
 
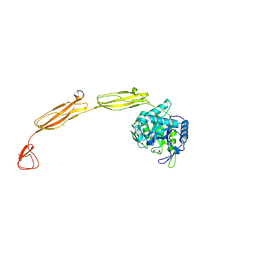 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HMD
 
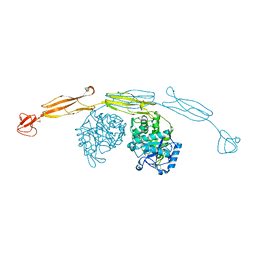 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HME
 
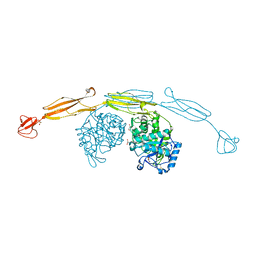 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMC
 
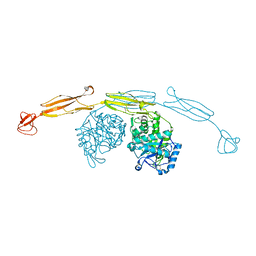 | | Crystal structure of cold-adapted chitinase from Moritella marina | | Descriptor: | Chitinase 60, GLYCEROL, SODIUM ION | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7BNH
 
 | |
7BNG
 
 | |
7BNI
 
 | |
7ZD8
 
 | | Crystal structure of the R24E mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD7
 
 | | Crystal structure of the R24E/E352T double mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD9
 
 | | Crystal structure of the E352T mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD4
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase soaked with Cu+ ions | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD0
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Cd2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD3
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Zn2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD1
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Hg2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD2
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Co2+ ions. | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
6ETE
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 5 | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, NICKEL (II) ION, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2017-10-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Crystal structure of KDM4D with tetrazolylhydrazide ligand NR128
To be published
|
|
6ETS
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
