6GKO
 
 | | Mouse thymidylate synthase cocrystallized with dUMP and soaked in phenolphthalein | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3,3-bis(4-hydroxyphenyl)-2-benzofuran-1-one, DIMETHYL SULFOXIDE, ... | | Authors: | Maj, P, Wilk, P, Jarmula, A, Weiss, M.S, Rode, W. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Thymidylate synthase fragment screening
To Be Published
|
|
6GYJ
 
 | | Mouse thymidylate synthase crystal soaked in phenolphthalein | | Descriptor: | 3,3-bis(4-hydroxyphenyl)-2-benzofuran-1-one, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Maj, P, Jarmula, A, Wilk, P, Weiss, M.S, Rode, W. | | Deposit date: | 2018-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thymidylate synthase fragment screening
To be published
|
|
6Y08
 
 | | Mouse thymidylate synthase cocrystallized with dUMP and soaked in sulfamethoxazole | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Sulfamethoxazole, Thymidylate synthase | | Authors: | Maj, P, Jarmula, A, Wilk, P, Weiss, M.S, Rode, W. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mouse thymidylate synthase cocrystallized with dUMP and soaked in sulfamethoxazole
To Be Published
|
|
6HE5
 
 | | 20S core particle of PAN-proteasomes | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta, Proteasome-activating nucleotidase | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HE9
 
 | | PAN-proteasome in state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.35 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HEA
 
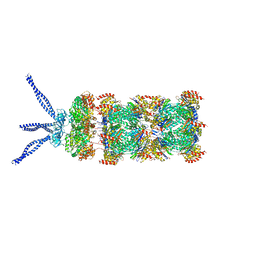 | | PAN-proteasome in state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.04 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HE4
 
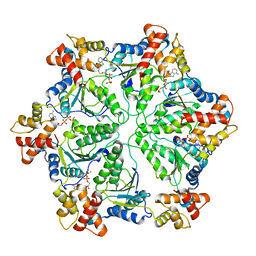 | | AAA-ATPase ring of PAN-proteasomes | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.85 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HE7
 
 | | 20S proteasome from Archaeoglobus fulgidus | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HE8
 
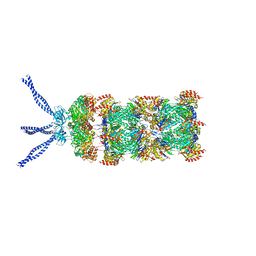 | | PAN-proteasome in state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.86 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HEC
 
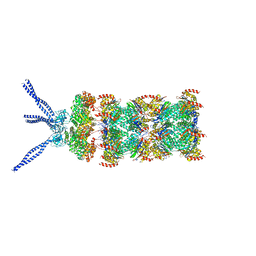 | | PAN-proteasome in state 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HED
 
 | | PAN-proteasome in state 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
9C68
 
 | |
9C69
 
 | |
9C6A
 
 | |
9C77
 
 | | cryoEM structure of CRISPR associated effector, CARF-Adenosine deaminase 1, Cad1, in cA4 bound form with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2024-06-10 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The CRISPR associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity
Cell(Cambridge,Mass.), 2024
|
|
9C6C
 
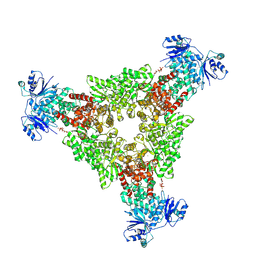 | | cryoEM structure of CRISPR associated effector, CARF-Adenosine deaminase 1, Cad1, in apo form with ATP (symmetric sites). | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The CRISPR associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity
Cell(Cambridge,Mass.), 2024
|
|
9C67
 
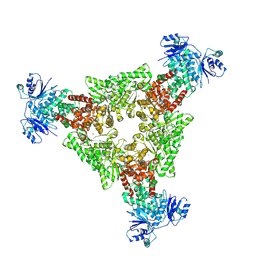 | |
9C6F
 
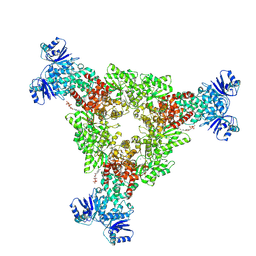 | | cryoEM structure of CRISPR associated effector, CARF-Adenosine deaminase 1, Cad1, in apo form with ATP (Asymmetric sites). | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The CRISPR associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity
Cell(Cambridge,Mass.), 2024
|
|
9CDB
 
 | | CryoEM structure of CRISPR associated effector, CARF-Adenosine deaminase 1, Cad1, in cA6 (partial density) bound form with ATP (partial density). | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2024-06-24 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The CRISPR associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity
Cell(Cambridge,Mass.), 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
