2XA6
 
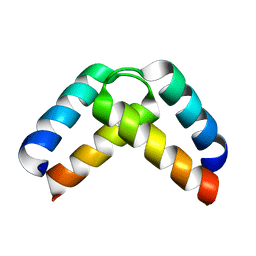 | | Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain | | Descriptor: | KH DOMAIN-CONTAINING,RNA-BINDING,SIGNAL TRANSDUCTION-ASSOCIATED PROTEIN 1 | | Authors: | Meyer, N.H, Tripsianes, K, Vincendeaux, M, Madl, T, Kateb, F, Brack-Werner, R, Sattler, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain.
J. Biol. Chem., 285, 2010
|
|
2JTW
 
 | |
2JMY
 
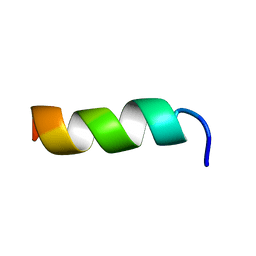 | | Solution structure of CM15 in DPC micelles | | Descriptor: | CM15 | | Authors: | Respondek, M, Madl, T, Goebl, C, Golser, R, Zangger, K. | | Deposit date: | 2006-12-13 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves
J.Am.Chem.Soc., 129, 2007
|
|
4BA8
 
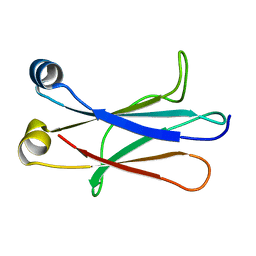 | | High resolution NMR structure of the C mu3 domain from IgM | | Descriptor: | IG MU CHAIN C REGION SECRETED FORM | | Authors: | Mueller, R, Kern, T, Graewert, M.A, Madl, T, Peschek, J, Groll, M, Sattler, M, Buchner, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-06-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High Resolution Structures of the Igm Fc Domains Reveal Principles of its Hexamer Formation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4A4E
 
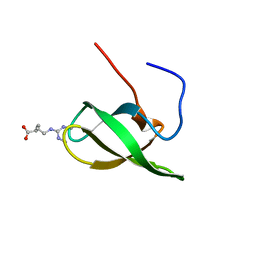 | | Solution structure of SMN Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL MOTOR NEURON PROTEIN | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A4H
 
 | | Solution structure of SPF30 Tudor domain in complex with asymmetrically dimethylated arginine | | Descriptor: | NG,NG-DIMETHYL-L-ARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A4F
 
 | | Solution structure of SPF30 Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4BXU
 
 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
2M64
 
 | | 1H, 13C and 15N Chemical Shift Assignments for Phl p 5a | | Descriptor: | Phlp5 | | Authors: | Goebl, C, Focke, M, Schrank, E, Madl, T, Kosol, S, Madritsch, C, Flicker, S, Valenta, R, Zangger, K, Tjandra, N. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexible IgE epitope-containing domains of Phl p 5 cause high allergenic activity.
J. Allergy Clin. Immunol., 140, 2017
|
|
4AKA
 
 | | IPSE alpha-1, an IgE-binding crystallin | | Descriptor: | IL-4-INDUCING PROTEIN | | Authors: | Meyer, N.H, Mayerhofer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Mueller-Dieckmann, J, Sattler, M, Scharmm, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-03-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Crystallin Fold in the Interleukin-4-Inducing Principle of Schistosoma Mansoni Eggs (Ipse/Alpha-1) Mediates Ige Binding for Antigen-Independent Basophil Activation
J.Biol.Chem., 290, 2015
|
|
4CZ3
 
 | | HP24wt derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
4CZ4
 
 | | HP24stab derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
