1O6Z
 
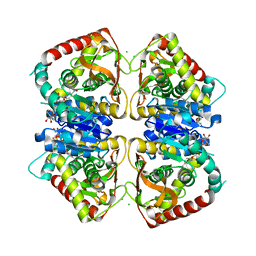 | | 1.95 A resolution structure of (R207S,R292S) mutant of malate dehydrogenase from the halophilic archaeon Haloarcula marismortui (holo form) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Ebel, C, Madern, D, Richard, S.B, Cosenza, L.W, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2002-10-22 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
9END
 
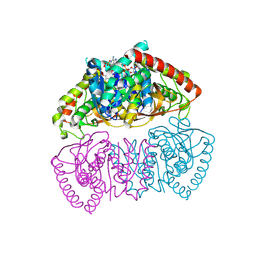 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 3 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Coquille, S, Roche, J, Girard, E, Madern, D. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To be published
|
|
3ZZN
 
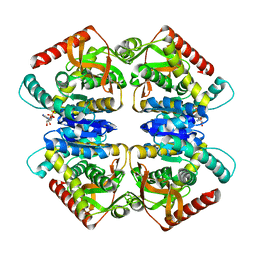 | | 5-Mutant (R79W, R151A, E279A, E299A,E313A) Lactate-Dehydrogenase from Thermus thermophillus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, LACTATE DEHYDROGENASE | | Authors: | Colletier, J.P, Mraihi, S, Madern, D. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sampling the conformational energy landscape of a hyperthermophilic protein by engineering key substitutions.
Mol. Biol. Evol., 29, 2012
|
|
7Q3X
 
 | |
1D3A
 
 | | CRYSTAL STRUCTURE OF THE WILD TYPE HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, HALOPHILIC MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-09-28 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
2X0R
 
 | | R207S, R292S Mutant of Malate Dehydrogenase from the Halophilic Archeon Haloarcula marismortui (HoloForm) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Irimia, A, Ebel, C, Vellieux, F.M.D, Richard, S.B, Cosenza, L.W, Zaccai, G, Madern, D. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
4BGV
 
 | |
4BGU
 
 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
4CL3
 
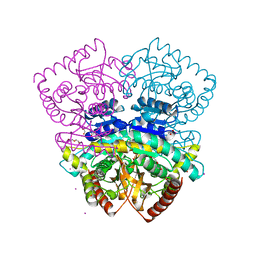 | | 1.70 A resolution structure of the malate dehydrogenase from Chloroflexus aurantiacus | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | An Experimental Point of View on Hydration/Solvation in Halophilic Proteins.
Front.Microbiol., 5, 2014
|
|
5MF2
 
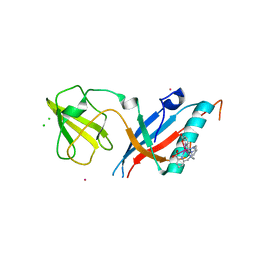 | | Bacteriophage T5 distal tail protein pb9 co-crystallized with Tb-Xo4 | | Descriptor: | CHLORIDE ION, Distal tail protein, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Lassalle, L, Arnaud, C.-A, Breyton, C, Madern, D, Coquelle, N, Maury, O, Girard, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallophore: a versatile lanthanide complex for protein crystallography combining nucleating effects, phasing properties, and luminescence.
Chem Sci, 8, 2017
|
|
8RS5
 
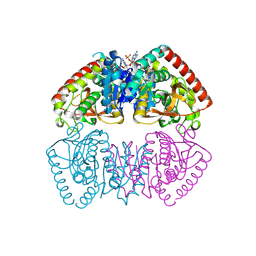 | |
8RWL
 
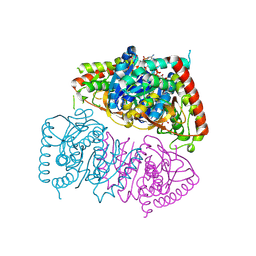 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Malate dehydrogenase, ... | | Authors: | Coquille, S, Roche, J, Engilberge, S, Girard, E, Madern, D. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To Be Published
|
|
8Q3C
 
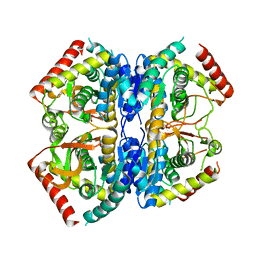 | | Structure of Selenomonas ruminantium lactate dehydrogenase I85R mutant | | Descriptor: | CHLORIDE ION, L-lactate dehydrogenase, NITRATE ION, ... | | Authors: | Bertrand, Q, Coquille, S, Iorio, A, Sterpone, F, Madern, D. | | Deposit date: | 2023-08-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
6QSS
 
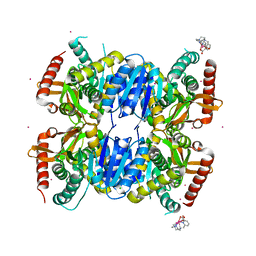 | | Crystal Structure of Ignicoccus islandicus malate dehydrogenase co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Roche, J, Girard, E, Madern, D. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (1.892 Å), X-RAY DIFFRACTION | | Cite: | The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
J.Struct.Biol., 208, 2019
|
|
6H9S
 
 | |
7NAY
 
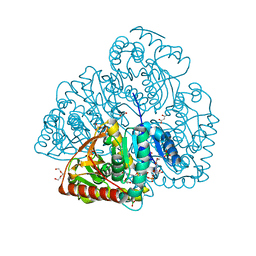 | | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, L-lactate dehydrogenase, ... | | Authors: | Bertrand, Q, Robin, A, Girard, E, Madern, D. | | Deposit date: | 2021-01-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
7AOB
 
 | | Crystal structure of Thermaerobacter marianensis malate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bertrand, Q, Lasalle, L, Girard, E, Madern, D. | | Deposit date: | 2020-10-14 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Thermaerobacter marianensis malate dehydrogenase
To Be Published
|
|
3NEP
 
 | |
2X0I
 
 | | 2.9 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.-K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
2X0J
 
 | | 2.8 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH ETHENO-NAD | | Descriptor: | ETHENO-NAD, MALATE DEHYDROGENASE, SULFATE ION | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
2J5R
 
 | | 2.25 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after second radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5Q
 
 | | 2.15 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after first radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5K
 
 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8AB2
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
