3EZ2
 
 | | Partition protein-ADP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Schumacher, M.A, Dunham, T.D, Xu, W, Funnell, B. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
4GCK
 
 | | structure of no-dna complex | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*C)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GFL
 
 | | NO mechanism, slma | | Descriptor: | Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1PNR
 
 | | PURINE REPRESSOR-HYPOXANTHINE-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1995-03-29 | | Release date: | 1995-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
1DBQ
 
 | | DNA-BINDING REGULATORY PROTEIN | | Descriptor: | MAGNESIUM ION, PURINE REPRESSOR | | Authors: | Schumacher, M.A, Choi, K.Y, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of corepressor-mediated specific DNA binding by the purine repressor.
Cell(Cambridge,Mass.), 83, 1995
|
|
1ZX4
 
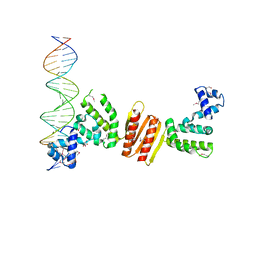 | | Structure of ParB bound to DNA | | Descriptor: | CITRIC ACID, Plasmid Partition par B protein, parS-small DNA centromere site | | Authors: | Schumacher, M.A, Funnell, B.E. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structures of ParB bound to DNA reveal mechanism of partition complex formation.
Nature, 438, 2005
|
|
1DBR
 
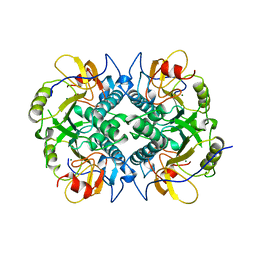 | | HYPOXANTHINE GUANINE XANTHINE | | Descriptor: | HYPOXANTHINE GUANINE XANTHINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Schumacher, M.A, Carter, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Toxoplasma gondii HGXPRTase reveal the catalytic role of a long flexible loop.
Nat.Struct.Biol., 3, 1996
|
|
4R4E
 
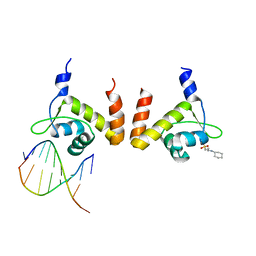 | | Structure of GlnR-DNA complex | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DNA (5'-D(*AP*TP*TP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*TP*A)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
3Q5W
 
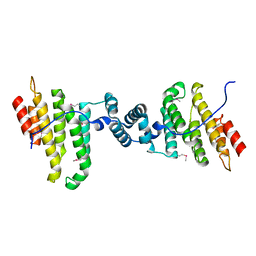 | |
3EZF
 
 | | Partition Protein | | Descriptor: | ParA, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
4GCT
 
 | | structure of No factor protein-DNA complex | | Descriptor: | DNA (5'-D(*TP*TP*AP*CP*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*CP*GP*TP*AP*A)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GFK
 
 | | structures of NO factors | | Descriptor: | Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1Q88
 
 | |
4FE4
 
 | | Crystal structure of apo E. coli XylR | | Descriptor: | Xylose operon regulatory protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2012-05-29 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: an AraC DNA-binding family member with a LacI/GalR ligand-binding domain.
Nucleic Acids Res., 41, 2013
|
|
3EZ6
 
 | | Structure of parA-ADP complex:tetragonal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partition protein A | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
1JJX
 
 | | Solution Structure of Recombinant Human Brain-type Fatty acid Binding Protein | | Descriptor: | BRAIN-TYPE FATTY ACID BINDING PROTEIN | | Authors: | Rademacher, M, Zimmerman, A.W, Rueterjans, H, Veerkamp, J.H, Luecke, C. | | Deposit date: | 2001-07-10 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of fatty acid-binding protein from human brain.
Mol.Cell.Biochem., 239, 2002
|
|
3EZ7
 
 | |
3EZ9
 
 | | Partition Protein | | Descriptor: | MAGNESIUM ION, ParA | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
1DH3
 
 | |
4R22
 
 | | TnrA-DNA complex | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), HTH-type transcriptional regulator TnrA | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4R24
 
 | | Complete dissection of B. subtilis nitrogen homeostatic circuitry | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), HTH-type transcriptional regulator TnrA | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
1Q87
 
 | |
1Q89
 
 | |
7NWW
 
 | | CspA-27 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-03-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIF
 
 | | CspA-27 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
