6TH2
 
 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
2VEK
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | 3-(BUTYLSULPHONYL)-PROPANOIC ACID, CITRIC ACID, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
6T5Z
 
 | | Crystal structure of an AA10 LPMO from Photorhabdus luminescens | | Descriptor: | COPPER (II) ION, Chitin-binding type-4 domain-containing protein | | Authors: | Munzone, A, El Kerdi, B, Reglier, M, Royant, A, Simaan, A.J, Decroos, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.60000312 Å) | | Cite: | Characterization of a bacterial copper-dependent lytic polysaccharide monooxygenase with an unusual second coordination sphere.
Febs J., 287, 2020
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
6T2O
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16 | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
6GKF
 
 | |
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6GVL
 
 | | Second pair of Fibronectin type III domains of integrin beta4 bound to the bullous pemphigoid antigen BP230 (BPAG1e) | | Descriptor: | Dystonin, Integrin beta-4 | | Authors: | Manso, J.A, Gomez-Hernandez, M, Alonso-Garcia, N, de Pereda, J.M. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Integrin alpha 6 beta 4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure, 27, 2019
|
|
6K6K
 
 | | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens
To Be Published
|
|
8FIV
 
 | |
6GMU
 
 | | Serum paraoxonase-1 by directed evolution with the L69G/H134R/F222S/T332S mutations | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2018-05-28 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzyme Evolution: An Epistatic Ratchet versus a Smooth Reversible Transition.
Mol.Biol.Evol., 37, 2020
|
|
8FIW
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor Jun10221 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide, N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors.
Eur.J.Med.Chem., 259, 2023
|
|
6JT6
 
 | | Crystal structure of cytochrome b domain of Pyranose Dehydrogenase from Coprinopsis cinerea | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | Deposit date: | 2019-04-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
6GO3
 
 | | TdT chimera (Loop1 of pol mu) - apoenzyme | | Descriptor: | DNA nucleotidylexotransferase,DNA-directed DNA/RNA polymerase mu,DNA nucleotidylexotransferase | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
2V8F
 
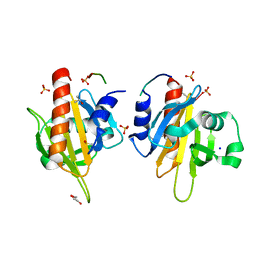 | | Mouse Profilin IIa in complex with a double repeat from the FH1 domain of mDia1 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Kursula, I, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
6TAI
 
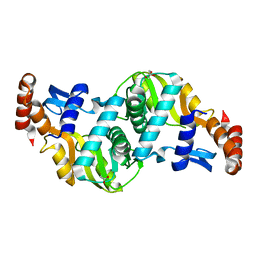 | | Crystal structure of Escherichia coli Orotate Phosphoribosyltransferase with an empty active site at 1.55 Angstrom resolution | | Descriptor: | ACETATE ION, GLYCEROL, Orotate phosphoribosyltransferase | | Authors: | Navas-Yuste, S, Lopez-Estepa, M, Gomez, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Elucidating the Catalytic Reaction Mechanism of Orotate Phosphoribosyltransferase by Means of X-ray Crystallography and Computational Simulations
Acs Catalysis, 10, 2020
|
|
6ZDB
 
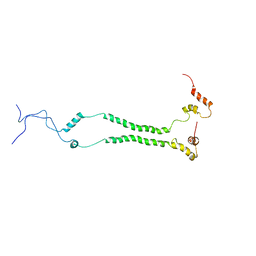 | | NMR structural analysis of yeast Cox13 reveals its C-terminus in interaction with ATP | | Descriptor: | Cytochrome c oxidase subunit 13, mitochondrial | | Authors: | Shu, Z, Pontus, P, Peter, B, Lena, M, Pia, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of the yeast cytochrome c oxidase subunit Cox13 and its interaction with ATP.
Bmc Biol., 19, 2021
|
|
1W0P
 
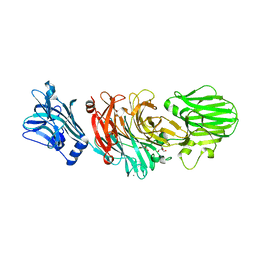 | | Vibrio cholerae sialidase with alpha-2,6-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Moustafa, I, Connaris, H, Taylor, M, Zaitsev, V, Wilson, J.C, Kiefel, M.J, von-Itzstein, M, Taylor, G. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sialic Acid Recognition by Vibrio Cholerae Neuraminidase.
J.Biol.Chem., 279, 2004
|
|
5LTN
 
 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease complexed with DPBA | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of Lymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
Iucrj, 5, 2018
|
|
2V98
 
 | | Structure of the complex of TcAChE with 1-(2-nitrophenyl)-2,2,2- trifluoroethyl-arsenocholine after a 9 seconds annealing to room temperature, during the first 5 seconds of which laser irradiation at 266nm took place | | Descriptor: | 1-(2-nitrophenyl)-2,2,2-trifluoroethyl]-arsenocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-22 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5LTX
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-MET | | Descriptor: | ACETATE ION, Chemotaxis protein, FORMIC ACID, ... | | Authors: | Gavira, J.A, Rico-Gimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
8DS6
 
 | |
8DRH
 
 | | HIGH RESOLUTION NMR STRUCTURE OF THE D(GCGTCAGG)R(CCUGACGC) HYBRID, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*CP*AP*GP*G)-3'), RNA (5'-R(*CP*CP*UP*GP*AP*CP*GP*C)-3') | | Authors: | Bachelin, M, Hessler, G, Kurz, G, Hacia, J.G, Dervan, P.B, Kessler, H. | | Deposit date: | 1997-10-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Nat.Struct.Biol., 5, 1998
|
|
